Biology:Epstein–Barr virus
| Human gammaherpesvirus 4 | |
|---|---|
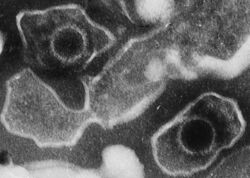
| |
| Electron micrograph of two Epstein–Barr virions (viral particles) showing round capsids loosely surrounded by the membrane envelope | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Peploviricota |
| Class: | Herviviricetes |
| Order: | Herpesvirales |
| Family: | Orthoherpesviridae |
| Genus: | Lymphocryptovirus |
| Species: | Human gammaherpesvirus 4
|
| Synonyms[1] | |
| |
The Epstein–Barr virus (EBV), formally called Human gammaherpesvirus 4, is one of the nine known human herpesvirus types in the herpes family, and is one of the most common viruses in humans. EBV is a double-stranded DNA virus.[2] Epstein-Barr virus (EBV) is the first identified oncogenic virus, which establishes permanent infection in humans. EBV causes infectious mononucleosis and is also tightly linked to many malignant diseases. Various vaccine formulations underwent testing in different animals or in humans. However, none of them was able to prevent EBV infection and no vaccine has been approved to date.[3]
The virus causes infectious mononucleosis ("mono" or "glandular fever"). It is also associated with various non-malignant, premalignant, and malignant Epstein–Barr virus-associated lymphoproliferative diseases such as Burkitt lymphoma, hemophagocytic lymphohistiocytosis,[4] and Hodgkin's lymphoma; non-lymphoid malignancies such as gastric cancer and nasopharyngeal carcinoma; and conditions associated with human immunodeficiency virus such as hairy leukoplakia and central nervous system lymphomas.[5][6] The virus is also associated with the childhood disorders of Alice in Wonderland syndrome[7] and acute cerebellar ataxia[8] and, by some evidence, higher risks of developing certain autoimmune diseases,[9] especially dermatomyositis, systemic lupus erythematosus, rheumatoid arthritis, and Sjögren's syndrome.[10][11] About 200,000 cancer cases globally per year are thought to be attributable to EBV.[12][13] In 2022, a large study (population of 10 million over 20 years) suggested EBV as the leading cause of multiple sclerosis, with a recent EBV infection causing a 32-fold increase in the risk of developing multiple sclerosis.[14][15][16][17][18]
Infection with EBV occurs by the oral transfer of saliva[19] and genital secretions. Most people become infected with EBV and gain adaptive immunity. In the United States, about half of all five-year-old children and about 90% of adults have evidence of previous infection.[20] Infants become susceptible to EBV as soon as maternal antibody protection disappears. Many children who become infected with EBV display no symptoms or the symptoms are indistinguishable from the other mild, brief illnesses of childhood.[21] When infection occurs during adolescence or young adulthood, it causes infectious mononucleosis 35 to 50% of the time.[22]
EBV infects B cells of the immune system and epithelial cells. Once EBV's initial lytic infection is brought under control, EBV latency persists in the individual's memory B cells for the rest of their life.[19][23][24]
Virology
Structure and genome
The virus is about 122–180 nm in diameter and is composed of a double helix of deoxyribonucleic acid (DNA) which contains about 172,000 base pairs encoding 85 genes.[19] The DNA is surrounded by a protein nucleocapsid, which is surrounded by a tegument made of protein, which in turn is surrounded by an envelope containing both lipids and surface projections of glycoproteins, which are essential to infection of the host cell.[25] In July 2020, a team of researchers reported the first complete atomic model of the nucleocapsid of the virus. This "first complete atomic model [includes] the icosahedral capsid, the capsid-associated tegument complex (CATC) and the dodecameric portal--the viral genome translocation apparatus." [26][27]
Tropism
The term viral tropism refers to which cell types that EBV infects. EBV can infect different cell types, including B cells and epithelial cells.[28]
The viral three-part glycoprotein complexes of gHgL gp42 mediate B cell membrane fusion; although the two-part complexes of gHgL mediate epithelial cell membrane fusion. EBV that are made in the B cells have low numbers of gHgLgp42 complexes, because these three-part complexes interact with Human-leukocyte-antigen class II molecules present in B cells in the endoplasmic reticulum and are degraded. In contrast, EBV from epithelial cells are rich in the three-part complexes because these cells do not normally contain HLA class II molecules. As a consequence, EBV made from B cells are more infectious to epithelial cells, and EBV made from epithelial cells are more infectious to B cells. Viruses lacking the gp42 portion are able to bind to human B cells, but unable to infect.[29]
Replication cycle
Entry to the cell
EBV can infect both B cells and epithelial cells. The mechanisms for entering these two cells are different.
To enter B cells, viral glycoprotein gp350 binds to cellular receptor CD21 (also known as CR2).[30] Then, viral glycoprotein gp42 interacts with cellular MHC class II molecules. This triggers fusion of the viral envelope with the cell membrane, allowing EBV to enter the B cell.[25] Human CD35, also known as complement receptor 1 (CR1), is an additional attachment factor for gp350 / 220, and can provide a route for entry of EBV into CD21-negative cells, including immature B-cells. EBV infection downregulates expression of CD35.[31]
To enter epithelial cells, viral protein BMRF-2 interacts with cellular β1 integrins. Then, viral protein gH/gL interacts with cellular αvβ6/αvβ8 integrins. This triggers fusion of the viral envelope with the epithelial cell membrane, allowing EBV to enter the epithelial cell.[25] Unlike B-cell entry, epithelial-cell entry is actually impeded by viral glycoprotein gp42.[30]
Once EBV enters the cell, the viral capsid dissolves and the viral genome is transported to the cell nucleus.[citation needed]
Lytic replication
The lytic cycle, or productive infection, results in the production of infectious virions. EBV can undergo lytic replication in both B cells and epithelial cells. In B cells, lytic replication normally only takes place after reactivation from latency. In epithelial cells, lytic replication often directly follows viral entry.[25]
For lytic replication to occur, the viral genome must be linear. The latent EBV genome is circular, so it must linearize in the process of lytic reactivation. During lytic replication, viral DNA polymerase is responsible for copying the viral genome. This contrasts with latency, in which host-cell DNA polymerase copies the viral genome.[25]
Lytic gene products are produced in three consecutive stages: immediate-early, early, and late.[25] Immediate-early lytic gene products act as transactivators, enhancing the expression of later lytic genes. Immediate-early lytic gene products include BZLF1 (also known as Zta, EB1, associated with its product gene ZEBRA) and BRLF1 (associated with its product gene Rta).[25] Early lytic gene products have many more functions, such as replication, metabolism, and blockade of antigen processing. Early lytic gene products include BNLF2.[25] Finally, late lytic gene products tend to be proteins with structural roles, such as VCA, which forms the viral capsid. Other late lytic gene products, such as BCRF1, help EBV evade the immune system.[25]
EGCG, a polyphenol in green tea, has shown in a study to inhibit EBV spontaneous lytic infection at the DNA, gene transcription, and protein levels in a time- and dose-dependent manner; the expression of EBV lytic genes Zta, Rta, and early antigen complex EA-D (induced by Rta), however, the highly stable EBNA-1 gene found across all stages of EBV infection is unaffected.[32] Specific inhibitors (to the pathways) suggest that Ras/MEK/MAPK pathway contributes to EBV lytic infection though BZLF1 and PI3-K pathway through BRLF1, the latter completely abrogating the ability of a BRLF1 adenovirus vector to induce the lytic form of EBV infection.[32] Additionally, the activation of some genes but not others is being studied to determine just how to induce immune destruction of latently infected B cells by use of either TPA or sodium butyrate.[32]
Latency
Unlike lytic replication, latency does not result in production of virions.[25] Instead, the EBV genome circular DNA resides in the cell nucleus as an episome and is copied by cellular DNA polymerase.[25] It persists in the individual's memory B cells.[19][24] Epigenetic changes such as DNA methylation and cellular chromatin constituents, suppress the majority of the viral genes in latently infected cells.[33] Only a portion of EBV's genes are expressed, which support the latent state of the virus.[33][19][34] Latent EBV expresses its genes in one of three patterns, known as latency programs. EBV can latently persist within B cells and epithelial cells, but different latency programs are possible in the two types of cell.[citation needed]
EBV can exhibit one of three latency programs: Latency I, Latency II, or Latency III. Each latency program leads to the production of a limited, distinct set of viral proteins and viral RNAs.[35][36]
| Gene Expressed | EBNA-1 | EBNA-2 | EBNA-3A | EBNA-3B | EBNA-3C | EBNA-LP | LMP1 | LMP-2A | LMP-2B | EBER |
|---|---|---|---|---|---|---|---|---|---|---|
| Product | Protein | Protein | Protein | Protein | Protein | Protein | Protein | Protein | Protein | ncRNAs |
| Latency I | + | – | – | – | – | – | – | – | – | + |
| Latency II | + | – | – | – | – | + | + | + | + | + |
| Latency III | + | + | + | + | + | + | + | + | + | + |
Also, a program is postulated in which all viral protein expression is shut off (Latency 0).[37]
Within B cells, all three latency programs are possible.[19] EBV latency within B cells usually progresses from Latency III to Latency II to Latency I. Each stage of latency uniquely influences B cell behavior.[19] Upon infecting a resting naïve B cell, EBV enters Latency III. The set of proteins and RNAs produced in Latency III transforms the B cell into a proliferating blast (also known as B cell activation).[19][25] Later, the virus restricts its gene expression and enters Latency II. The more limited set of proteins and RNAs produced in Latency II induces the B cell to differentiate into a memory B cell.[19][25] Finally, EBV restricts gene expression even further and enters Latency I. Expression of EBNA-1 allows the EBV genome to replicate when the memory B cell divides.[19][25]
Within epithelial cells, only Latency II is possible.[38]
In primary infection, EBV replicates in oropharyngeal epithelial cells and establishes Latency III, II, and I infections in B lymphocytes. EBV latent infection of B lymphocytes is necessary for virus persistence, subsequent replication in epithelial cells, and release of infectious virus into saliva. EBV Latency III and II infections of B lymphocytes, Latency II infection of oral epithelial cells, and Latency II infection of NK- or T-cell can result in malignancies, marked by uniform EBV genome presence and gene expression.[39]
Reactivation
Latent EBV in B cells can be reactivated to switch to lytic replication. This is known to happen in vivo, but what triggers it is not known precisely. In vitro, latent EBV in B cells can be reactivated by stimulating the B cell receptor, so it is likely reactivation in vivo takes place after latently infected B cells respond to unrelated infections.[25]
Transformation of B lymphocytes
When EBV infects B cells in vitro, lymphoblastoid cell lines eventually emerge that are capable of indefinite growth. The growth transformation of these cell lines is the consequence of viral protein expression.[40]
EBNA-2, EBNA-3C, and LMP-1, are essential for transformation, whereas EBNA-LP and the EBERs are not.[41]
Following natural infection with EBV, the virus is thought to execute some or all of its repertoire of gene expression programs to establish a persistent infection. Given the initial absence of host immunity, the lytic cycle produces large numbers of virions to infect other (presumably) B-lymphocytes within the host.
The latent programs reprogram and subvert infected B-lymphocytes to proliferate and bring infected cells to the sites at which the virus presumably persists. Eventually, when host immunity develops, the virus persists by turning off most (or possibly all) of its genes and only occasionally reactivates and produces progeny virions. A balance is eventually struck between occasional viral reactivation and host immune surveillance removing cells that activate viral gene expression. The manipulation of the human body's epigenetics by EBV can alter the genome of the cell to leave oncogenic phenotypes.[42] As a result, the modification by the EBV increases the hosts likelihood of developing EBV related cancer.[43] EBV related cancers are unique in that they are frequent to making epigenetic changes but are less likely to mutate.[44]
The site of persistence of EBV may be bone marrow. EBV-positive patients who have had their own bone marrow replaced with bone marrow from an EBV-negative donor are found to be EBV-negative after transplantation.[45]
Latent antigens
All EBV nuclear proteins are produced by alternative splicing of a transcript starting at either the Cp or Wp promoters at the left end of the genome (in the conventional nomenclature). The genes are ordered EBNA-LP/EBNA-2/EBNA-3A/EBNA-3B/EBNA-3C/EBNA-1 within the genome.
The initiation codon of the EBNA-LP coding region is created by an alternate splice of the nuclear protein transcript. In the absence of this initiation codon, EBNA-2/EBNA-3A/EBNA-3B/EBNA-3C/EBNA-1 will be expressed depending on which of these genes is alternatively spliced into the transcript.
Protein/genes
| Protein/gene/antigen | Stage | Description |
|---|---|---|
| EBNA-1 | latent+lytic | EBNA-1 protein binds to a replication origin (oriP) within the viral genome and mediates replication and partitioning of the episome during division of the host cell. It is the only viral protein expressed during group I latency. |
| EBNA-2 | latent+lytic | EBNA-2 is the main viral transactivator. |
| EBNA-3 | latent+lytic | These genes also bind the host RBP-Jκ protein. |
| LMP-1 | latent | LMP-1 is a six-span transmembrane protein that is also essential for EBV-mediated growth transformation. |
| LMP-2 | latent | LMP-2A/LMP-2B are transmembrane proteins that act to block tyrosine kinase signaling. |
| EBER | latent | EBER-1/EBER-2 are small nuclear RNAs, which bind to certain nucleoprotein particles, enabling binding to PKR (dsRNA-dependent serin/threonin protein kinase), thus inhibiting its function. EBERs are by far the most abundant EBV products transcribed in EBV-infected cells. They are commonly used as targets for the detection of EBV in histological tissues.[46] ER-particles also induce the production of IL-10, which enhances growth and inhibits cytotoxic T cells. |
| v-snoRNA1 | latent | Epstein–Barr virus snoRNA1 is a box CD-snoRNA generated by the virus during latency. V-snoRNA1 may act as a miRNA-like precursor that is processed into 24 nucleotide sized RNA fragments that target the 3'UTR of viral DNA polymerase mRNA.[36] |
| ebv-sisRNA | latent | Ebv-sisRNA-1 is a stable intronic sequence RNA generated during latency program III. After the EBERs, it is the third-most abundant small RNA produced by the virus during this program.[47] |
| miRNAs | latent | EBV microRNAs are encoded by two transcripts, one set in the BART gene and one set near the BHRF1 cluster. The three BHRF1 pri-miRNAS (generating four miRNAs) are expressed during type III latency, whereas the large cluster of BART miRNAs (up to 20 miRNAs) are expressed highly during type II latency and only modestly during type I and II latency.[48] The previous reference also gives an account of the known functions of these miRNAs. |
| EBV-EA | lytic | early antigen |
| EBV-MA | lytic | membrane antigen |
| EBV-VCA | lytic | viral capsid antigen |
| EBV-AN | lytic | alkaline nuclease[49] |
Subtypes of EBV
EBV can be divided into two major types, EBV type 1 and EBV type 2. These two subtypes have different EBNA-3 genes. As a result, the two subtypes differ in their transforming capabilities and reactivation ability. Type 1 is dominant throughout most of the world, but the two types are equally prevalent in Africa. One can distinguish EBV type 1 from EBV type 2 by cutting the viral genome with a restriction enzyme and comparing the resulting digestion patterns by gel electrophoresis.[25]
Detection
Epstein–Barr virus-encoded small RNAs (EBERs) are by far the most abundant EBV products transcribed in cells infected by EBV. They are commonly used as targets for the detection of EBV in histological tissues.[46]
Role in disease
- See also Infectious mononucleosis and the other diseases listed in this section
EBV causes infectious mononucleosis.[50] Children infected with EBV have few symptoms or can appear asymptomatic, but when infection is delayed to adolescence or adulthood, it can cause fatigue, fever, inflamed throat, swollen lymph nodes in the neck, enlarged spleen, swollen liver, or rash.[20] Post-infectious chronic fatigue syndrome has also been associated with EBV infection.[51][52]
EBV has also been implicated in several other diseases, including Burkitt's lymphoma,[53] hemophagocytic lymphohistiocytosis,[54] Hodgkin's lymphoma,[55] stomach cancer,[12][56] nasopharyngeal carcinoma,[57] multiple sclerosis,[15][16][58][17] and lymphomatoid granulomatosis.[59]
Specifically, EBV infected B cells have been shown to reside within the brain lesions of multiple sclerosis patients,[17] and a 2022 study of 10 million soldiers' historical blood samples showed that "Individuals who were not infected with the Epstein-Barr virus virtually never get multiple sclerosis. It's only after Epstein-Barr virus infection that the risk of multiple sclerosis jumps up by over 30 fold", and that only EBV of many infections had such a clear connection with the disease.[60]
Additional diseases that have been linked to EBV include Gianotti–Crosti syndrome, erythema multiforme, acute genital ulcers, and oral hairy leukoplakia.[61] The viral infection is also associated with, and often contributes to the development of, a wide range of non-malignant lymphoproliferative diseases such as severe hypersensitivity mosquito bite allergy reactions,[62] Epstein-Barr virus-positive mucocutaneous ulcers, and hydroa vacciniforme as well as malignant lymphoproliferative diseases such as Epstein–Barr virus-positive Burkitt lymphoma,[63] Epstein–Barr virus-positive Hodgkin lymphoma,[64] and primary effusion lymphoma.[65]
The Epstein–Barr virus has been implicated in disorders related to alpha-synuclein aggregation (e.g. Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy).[66]
It has been found that EBNA1 may induce chromosomal breakage in the 11th chromosome, specifically in the 11q23 region between the FAM55D gene and FAM55B, which EBNA-1 appears to have a high affinity for due to its DNA-binding domain having an interest in a specific palindromic repeat in this section of the genome.[67] While the cause and exact mechanism for this is unknown, the byproduct results in errors and breakage of the chromosomal structure as cells stemming from the line of the tainted genome undergo mitosis. Since genes in this area have been implicated in leukemia and is home to a tumor suppressor gene that is modified or not present in most tumor gene expression, it's been hypothesized that breakage in this area is the main culprit behind the cancers that EBV increases the chance of. The breakage is also dose-dependent, a person with a latent infection will have less breakage than a person with a novel or reactivated infection since EBNA1 levels in the nucleus and nucleolus are higher during active attack of the body because of the constant replication and take-over of cells in the body.
History
The Epstein–Barr virus was named after M.A. Epstein, and Yvonne Barr, who discovered the virus together with Bert Achong.[68][69] In 1961, Epstein, a pathologist and expert electron microscopist, attended a lecture on "The commonest children's cancer in tropical Africa — a hitherto unrecognised syndrome" by D.P. Burkitt, a surgeon practicing in Uganda, in which Burkitt described the "endemic variant" (pediatric form) of the disease that now bears his name. In 1963, a specimen was sent from Uganda to Middlesex Hospital to be cultured. Virus particles were identified in the cultured cells, and the results were published in The Lancet in 1964 by Epstein, Achong, and Barr.[69][70] Cell lines were sent to Werner and Gertrude Henle at the Children's Hospital of Philadelphia who developed serological markers.[71] In 1967, a technician in their laboratory developed mononucleosis and they were able to compare a stored serum sample, showing that antibodies to the virus developed.[70][72][73] In 1968, they discovered that EBV can directly immortalize B cells after infection, mimicking some forms of EBV-related infections,[71] and confirmed the link between the virus and infectious mononucleosis.[74]
Research
As a relatively complex virus, EBV is not yet fully understood. Laboratories around the world continue to study the virus and develop new ways to treat the diseases it causes. One popular way of studying EBV in vitro is to use bacterial artificial chromosomes.[75] Epstein–Barr virus can be maintained and manipulated in the laboratory in continual latency (a property shared with Kaposi's sarcoma-associated herpesvirus, another of the eight human herpesviruses). Although many viruses are assumed to have this property during infection of their natural hosts, there is not an easily managed system for studying this part of the viral lifecycle. Genomic studies of EBV have been able to explore lytic reactivation and regulation of the latent viral episome.[76]
Although under active research, an Epstein–Barr virus vaccine is not yet available. The development of an effective vaccine could prevent up to 200,000 cancers globally per year.[12] The absence of effective animal models is an obstacle to development of prophylactic and therapeutic vaccines against EBV.[24]
Like other human herpesviruses Epstein-Barr might allow its own eradication via a course of the drug valaciclovir, but further research is needed to determine if eradication is actually achievable.[42] Antiviral agents act by inhibiting viral DNA replication, but there is little evidence that they are effective against Epstein–Barr virus. Moreover, they are expensive, risk causing resistance to antiviral agents, and (in 1% to 10% of cases) can cause unpleasant side effects.[43]
See also
- Epstein–Barr virus infection
- Epstein–Barr virus-associated lymphoproliferative diseases
- James Corson Niederman, the physician who proved how the Epstein–Barr virus is transmitted in infectious mononucleosis
References
- ↑ "ICTV Taxonomy history: Human gammaherpesvirus 4". https://ictv.global/taxonomy/taxondetails?taxnode_id=20181486.
- ↑ "Beyond cytomegalovirus and Epstein-Barr virus: A feview of viruses composing the blood virome of solid organ transplant and hematopoietic stem cell transplant recipients". Clinical Microbiology Reviews 33 (4): e00027-20. 2020. doi:10.1128/CMR.00027-20. PMID 32847820.
- ↑ Urgency and necessity of Epstein-Barr virus prophylactic vaccines. 2022. npj Vaccines. 7/1. L. Zhong, C. Krummenacher, W. Zhang, J. Hong, Q. Feng, Y. Chen, et al. doi: 10.1038/s41541-022-00587-6.
- ↑ "Epstein-Barr virus (EBV)-associated lymphoid proliferations, a 2018 update". Human Pathology 79: 18–41. September 2018. doi:10.1016/j.humpath.2018.05.020. PMID 29885408.
- ↑ "Spectrum of Epstein-Barr virus-related diseases: A pictorial review". Japanese Journal of Radiology 27 (1): 4–19. January 2009. doi:10.1007/s11604-008-0291-2. PMID 19373526.
- ↑ "Oral manifestations in the era of HAART". Journal of the National Medical Association 95 (2, Supplement 2): 21S–32S. February 2003. PMID 12656429.
- ↑ "Alice in Wonderland Syndrome: A clinical and pathophysiological review". BioMed Research International 2016: 8243145. 2016. doi:10.1155/2016/8243145. PMID 28116304.
- ↑ "Post-infectious acute cerebellar ataxia in children". Clinical Pediatrics 42 (7): 581–584. September 2003. doi:10.1177/000992280304200702. PMID 14552515.
- ↑ "Epstein-Barr virus in autoimmune diseases". Best Practice & Research: Clinical Rheumatology 22 (5): 883–896. October 2008. doi:10.1016/j.berh.2008.09.007. PMID 19028369.
- ↑ "Autoimmune disease: A role for new anti-viral therapies?". Autoimmunity Reviews 11 (2): 88–97. December 2011. doi:10.1016/j.autrev.2011.08.005. PMID 21871974.
- ↑ "CD8+ T cell deficiency, Epstein-Barr virus infection, vitamin D deficiency, and steps to autoimmunity: A unifying hypothesis". Autoimmune Diseases 2012: 189096. 2012. doi:10.1155/2012/189096. PMID 22312480.
- ↑ 12.0 12.1 12.2 "Developing a vaccine for the Epstein–Barr virus could prevent up to 200,000 cancers globally say experts". Cancer Research UK (Press release). 2014-03-24. Archived from the original on 2017-03-19.
- ↑ "Global and regional incidence, mortality and disability-adjusted life-years for Epstein-Barr virus-attributable malignancies, 1990-2017". BMJ Open 10 (8): e037505. August 2020. doi:10.1136/bmjopen-2020-037505. PMID 32868361.
- ↑ "Epstein-Barr virus and multiple sclerosis". Science 375 (6578): 264–265. January 2022. doi:10.1126/science.abm7930. PMID 35025606. Bibcode: 2022Sci...375..264R.
- ↑ 15.0 15.1 "Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis". Science (American Association for the Advancement of Science (AAAS)) 375 (6578): 296–301. January 2022. doi:10.1126/science.abj8222. PMID 35025605. Bibcode: 2022Sci...375..296B. Related non-technical article: "Can we vaccinate against Epstein-Barr, the virus you didn't know you had?". The Observer. 20 March 2022. https://www.theguardian.com/science/2022/mar/20/can-we-vaccinate-against-epstein-barr-virus-multiple-sclerosis-cancer.
- ↑ 16.0 16.1 "Epstein-barr virus infection and multiple sclerosis: a review". Journal of Neuroimmune Pharmacology 5 (3): 271–277. September 2010. doi:10.1007/s11481-010-9201-3. PMID 20369303.
- ↑ 17.0 17.1 17.2 "Molecular signature of Epstein-Barr virus infection in MS brain lesions". Neurology 5 (4): e466. July 2018. doi:10.1212/NXI.0000000000000466. PMID 29892607.
- ↑ "Epstein-Barr virus is present in the brain of most cases of multiple sclerosis and may engage more than just B cells". PLoS One 13 (2): e0192109. 2018. doi:10.1371/journal.pone.0192109. PMID 29394264. Bibcode: 2018PLoSO..1392109H.
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 19.6 19.7 19.8 19.9 "Reactivation of Epstein-Barr virus from latency". Reviews in Medical Virology 15 (3): 149–156. November 2004. doi:10.1002/rmv.456. PMID 15546128.
- ↑ 20.0 20.1 "About 90% of adults have antibodies that show that they have a current or past EBV infection". US CDC. 28 September 2020. https://www.cdc.gov/epstein-barr/about-ebv.html.
- ↑ "NIH conference. Epstein-Barr virus infections: Biology, pathogenesis, and management". Annals of Internal Medicine 118 (1): 45–58. January 1993. doi:10.7326/0003-4819-118-1-199301010-00009. PMID 8380053.
- ↑ "Epstein–Barr virus and infectious Mononucleosis". U.S. Centers for Disease Control and Prevention (CDC). https://www.cdc.gov/ncidod/diseases/ebv.htm.
- ↑ "Is EBV persistence in vivo a model for B cell homeostasis?". Immunity 5 (2): 173–179. August 1996. doi:10.1016/s1074-7613(00)80493-8. PMID 8769480.
- ↑ 24.0 24.1 24.2 "Main Targets of Interest for the Development of a Prophylactic or Therapeutic Epstein-Barr Virus Vaccine". Frontiers in Microbiology 12: 701611. 2021. doi:10.3389/fmicb.2021.701611. PMID 34239514.
- ↑ 25.00 25.01 25.02 25.03 25.04 25.05 25.06 25.07 25.08 25.09 25.10 25.11 25.12 25.13 25.14 25.15 "Progress and problems in understanding and managing primary Epstein-Barr virus infections". Clinical Microbiology Reviews 24 (1): 193–209. January 2011. doi:10.1128/CMR.00044-10. PMID 21233512.
- ↑ Jia L (July 17, 2020). "Scientists uncover first atomic structure of Epstein-Bar virus nucleocapsid". phys.org (Press release). Retrieved October 4, 2020.
- ↑ "CryoEM structure of the tegumented capsid of Epstein-Barr virus". Cell Research 30 (10): 873–884. October 2020. doi:10.1038/s41422-020-0363-0. PMID 32620850.
- ↑ "Epstein Barr virus entry; kissing and conjugation". Current Opinion in Virology 4: 78–84. February 2014. doi:10.1016/j.coviro.2013.12.001. PMID 24553068.
- ↑ "Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect". Journal of Virology 72 (1): 158–163. January 1998. doi:10.1128/jvi.72.1.158-163.1998. PMID 9420211.
- ↑ 30.0 30.1 "Entrez gene: CR2 complement component (3d/Epstein Barr virus) receptor 2". https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=ShowDetailView&TermToSearch=1380.
- ↑ "Human complement receptor type 1 / CD35 is an Epstein-Barr Virus receptor". Cell Reports 3 (2): 371–85. February 2013. doi:10.1016/j.celrep.2013.01.023. PMID 23416052.
- ↑ 32.0 32.1 32.2 "(-)-Epigallocatechin-3-gallate inhibition of Epstein-Barr virus spontaneous lytic infection involves ERK1/2 and PI3-K/Akt signaling in EBV-positive cells". Carcinogenesis 34 (3): 627–637. March 2013. doi:10.1093/carcin/bgs364. PMID 23180656.
- ↑ 33.0 33.1 "Epigenetic lifestyle of Epstein-Barr virus". Seminars in Immunopathology 42 (2): 131–142. April 2020. doi:10.1007/s00281-020-00792-2. PMID 32232535.
- ↑ "Epstein-Barr virus and the B cell: That's all it takes". Trends in Microbiology 4 (5): 204–208. May 1996. doi:10.1016/s0966-842x(96)90020-7. PMID 8727601.
- ↑ "Epstein-Barr virus and virus human protein interaction maps". Proceedings of the National Academy of Sciences of the United States of America 104 (18): 7606–11. May 2007. doi:10.1073/pnas.0702332104. PMID 17446270. Bibcode: 2007PNAS..104.7606C. The nomenclature used here is that of Kieff. Other laboratories use different nomenclatures.
- ↑ 36.0 36.1 "Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome". PLOS Pathogens 5 (8): e1000547. August 2009. doi:10.1371/journal.ppat.1000547. PMID 19680535.
- ↑ "Microbiome and malignancy". Cell Host & Microbe 10 (4): 324–335. October 2011. doi:10.1016/j.chom.2011.10.003. PMID 22018233.
- ↑ "Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells". Journal of Virology 87 (15): 8606–8623. August 2013. doi:10.1128/JVI.00805-13. PMID 23720728.
- ↑ Epstein–Barr Virus: Latency and Transformation. Caister Academic Press. 2010. ISBN 978-1-904455-62-2.
- ↑ "Latency and lytic replication in Epstein-Barr virus-associated oncogenesis". Nature Reviews Microbiology 17 (11): 691–700. November 2019. doi:10.1038/s41579-019-0249-7. PMID 31477887. https://www.zora.uzh.ch/id/eprint/183065/9/latecny_and_lytic_replication_in_the_oncogenesis_of_the_epstein-barr_virus_final_manuscript.pdf.
- ↑ "Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells". Nature 313 (6005): 812–815. 1985. doi:10.1038/313812a0. PMID 2983224. Bibcode: 1985Natur.313..812Y.
- ↑ 42.0 42.1 "Long-term administration of valacyclovir reduces the number of Epstein-Barr virus (EBV)-infected B cells but not the number of EBV DNA copies per B cell in healthy volunteers". Journal of Virology 83 (22): 11857–11861. November 2009. doi:10.1128/JVI.01005-09. PMID 19740997.
- ↑ 43.0 43.1 "Antiviral agents for infectious mononucleosis (glandular fever)". The Cochrane Database of Systematic Reviews 2016 (12): CD011487. 2016. doi:10.1002/14651858.CD011487.pub2. PMID 27933614.
- ↑ "Epstein-Barr virus: A master epigenetic manipulator". Current Opinion in Virology 26: 74–80. October 2017. doi:10.1016/j.coviro.2017.07.017. PMID 28780440.
- ↑ "Eradication of Epstein-Barr virus by allogeneic bone marrow transplantation: implications for sites of viral latency". Proceedings of the National Academy of Sciences of the United States of America 85 (22): 8693–8696. November 1988. doi:10.1073/pnas.85.22.8693. PMID 2847171. Bibcode: 1988PNAS...85.8693G.
- ↑ 46.0 46.1 "Epstein Barr virus (EBV) encoded small RNAs: Targets for detection by in situ hybridisation with oligonucleotide probes". Journal of Clinical Pathology 45 (7): 616–620. July 1992. doi:10.1136/jcp.45.7.616. PMID 1325480.
- ↑ "Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA". BMC Genomics 14: 543. August 2013. doi:10.1186/1471-2164-14-543. PMID 23937650.
- ↑ "The role of microRNAs in Epstein-Barr virus latency and lytic reactivation". Microbes Infect 13 (14,15): 1156–1167. December 2011. doi:10.1016/j.micinf.2011.07.007. PMID 21835261.
- ↑ "A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities". Journal of Molecular Biology 391 (4): 717–728. August 2009. doi:10.1016/j.jmb.2009.06.034. PMID 19538972.
- ↑ "Benign lymphadenopathies". Modern Pathology 26 (Supplement 1): S88–S96. January 2013. doi:10.1038/modpathol.2012.176. PMID 23281438.
- ↑ "Chronic fatigue syndrome: A manifestation of Epstein-Barr virus infection?". Current Clinical Topics in Infectious Diseases 9: 126–146. 1988. PMID 2855828.
- ↑ "Long COVID or post-acute sequelae of COVID-19 (PASC): An overview of biological factors that may contribute to persistent symptoms". Frontiers in Microbiology 12: 698169. 23 June 2021. doi:10.3389/fmicb.2021.698169. PMID 34248921.
- ↑ "The role of EBV in the pathogenesis of Burkitt's Lymphoma: an Italian hospital based survey". Infectious Agents and Cancer 9 (1): 34. 2014. doi:10.1186/1750-9378-9-34. PMID 25364378.
- ↑ "Epstein–Barr virus and hemophagocytic lymphohistiocytosis". Frontiers in Immunology 8: 1902. 2017. doi:10.3389/fimmu.2017.01902. PMID 29358936.
- ↑ "Epstein-Barr virus-associated Hodgkin's lymphoma". British Journal of Haematology 125 (3): 267–281. May 2004. doi:10.1111/j.1365-2141.2004.04902.x. PMID 15086409.
- ↑ "Epigenetic dysregulation in Epstein-Barr virus-associated gastric carcinoma: Disease and treatments". World Journal of Gastroenterology 20 (21): 6448–6456. June 2014. doi:10.3748/wjg.v20.i21.6448. PMID 24914366.
- ↑ "Human papillomavirus and Epstein-Barr virus in nasopharyngeal carcinoma in a low-incidence population". Head & Neck 36 (4): 511–516. April 2014. doi:10.1002/hed.23318. PMID 23780921.
- ↑ "Epstein-Barr virus genetic variants are associated with multiple sclerosis". Neurology 84 (13): 1362–1368. March 2015. doi:10.1212/WNL.0000000000001420. PMID 25740864.
- ↑ "Lymphomatoid granulomatosis: A practical review for pathologists dealing with this rare pulmonary lymphoproliferative process". Pathologica 105 (4): 111–116. August 2013. PMID 24466760.
- ↑ "Is a virus we all have causing multiple sclerosis?". 13 April 2022. https://www.bbc.com/news/health-61042598.
- ↑ "Epstein-Barr virus and skin manifestations in childhood". International Journal of Dermatology 52 (10): 1177–1184. October 2013. doi:10.1111/j.1365-4632.2012.05855.x. PMID 24073903.
- ↑ "Primary EBV infection and hypersensitivity to mosquito bites: a case report". Virologica Sinica 31 (6): 517–520. December 2016. doi:10.1007/s12250-016-3868-4. PMID 27900557.
- ↑ "The presence of Epstein–Barr virus significantly impacts the transcriptional profile in immunodeficiency-associated Burkitt lymphoma". Frontiers in Microbiology 6: 556. 2015. doi:10.3389/fmicb.2015.00556. PMID 26113842.
- ↑ "Epstein-Barr virus-associated lymphomas". Philosophical Transactions of the Royal Society of London Series B, Biological Sciences 372 (1732): 20160271. October 2017. doi:10.1098/rstb.2016.0271. PMID 28893938.
- ↑ "Primary effusion lymphoma: Current concepts and management". Current Opinion in Pulmonary Medicine 23 (4): 365–370. July 2017. doi:10.1097/MCP.0000000000000384. PMID 28399009.
- ↑ "Monoclonal antibodies against Epstein-Barr virus cross-react with alpha-synuclein in human brain". Neurology 55 (9): 1398–1401. November 2000. doi:10.1212/WNL.55.9.1398. PMID 11087792.
- ↑ Frappier, Lori (April 2023). "Epstein–Barr virus is an agent of genomic instability". Nature 616 (7957): 441–442. doi:10.1038/d41586-023-00936-y. PMID 37045952. Bibcode: 2023Natur.616..441F. https://www.nature.com/articles/d41586-023-00936-y.
- ↑ "Cancer virus discovery helped by delayed flight". 6 April 2014. https://www.bbc.co.uk/news/health-26857610.
- ↑ 69.0 69.1 "Virus Particles in Cultured Lymphoblasts from Burkitt's Lymphoma". Lancet 1 (7335): 702–703. March 1964. doi:10.1016/s0140-6736(64)91524-7. PMID 14107961.
- ↑ 70.0 70.1 "1. The origins of EBV research: Discovery and characterization of the virus". Epstein–Barr Virus. Wymondham, UK: Caister Academic Press. 2005. pp. 1–14. ISBN 978-1-904455-03-5. https://books.google.com/books?id=TRO-wXto8hcC. Retrieved September 18, 2010.
- ↑ 71.0 71.1 "Epidemiologic aspects of Epstein-Barr virus (EBV)-associated diseases". Annals of the New York Academy of Sciences 354: 326–331. 1980. doi:10.1111/j.1749-6632.1980.tb27975.x. PMID 6261650.
- ↑ Epstein–Barr Virus. Horizon Scientific Press. 2005. p. 18. ISBN 978-1-904455-03-5. https://books.google.com/books?id=TRO-wXto8hcC. Retrieved 3 June 2012.
- ↑ "Epstein–Barr Virus". New England Journal of Medicine 355 (25): 2708–2709. 21 December 2006. doi:10.1056/NEJMbkrev39523.
- ↑ Desk Encyclopedia of Human and Medical Virology. Boston, MA: Academic Press. 2009. pp. 532–533.
- ↑ "Contribution of viral recombinants to the study of the immune response against the Epstein-Barr virus". Seminars in Cancer Biology 18 (6): 409–415. December 2008. doi:10.1016/j.semcancer.2008.09.001. PMID 18938248.
- ↑ "An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions". Cell Host & Microbe 12 (2): 233–245. August 2012. doi:10.1016/j.chom.2012.06.008. PMID 22901543.
Further reading
- "The puzzling virus that infects almost everyone". The Atlantic. 3 Mar 2022. https://www.theatlantic.com/science/archive/2022/03/epstein-barr-virus-mono-cancer-research/623881/.
External links
Wikidata ☰ Q24808752 entry
 |



