Biology:Gammaherpesvirinae
| Gammaherpesvirinae | |
|---|---|
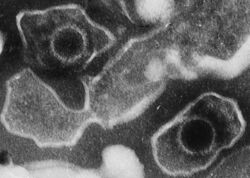
| |
| Electron micrograph of two Human gammaherpesvirus 4 virions (viral particles) showing round capsids loosely surrounded by the membrane envelope | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Peploviricota |
| Class: | Herviviricetes |
| Order: | Herpesvirales |
| Family: | Orthoherpesviridae |
| Subfamily: | Gammaherpesvirinae |
| Genera | |
|
See text | |
Gammaherpesvirinae is a subfamily of viruses in the order Herpesvirales and in the family Herpesviridae. Viruses in Gammaherpesvirinae are distinguished by reproducing at a more variable rate than other subfamilies of Herpesviridae. Mammals serve as natural hosts. There are 43 species in this subfamily, divided among 7 genera with three species unassigned to a genus. Diseases associated with this subfamily include: HHV-4: infectious mononucleosis. HHV-8: Kaposi's sarcoma.[1][2]
Taxonomy
Herpesviruses represent a group of double-stranded DNA viruses distributed widely within the animal kingdom. The family Herpesviridae, which contains eight viruses that infect humans, is the most extensively studied group within this order and comprises three subfamilies, namely Alphaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae.
Within the Gammaherpesvirinae there are a number of unclassified viruses including Cynomys herpesvirus 1 (CynGHV-1)[3] Elephantid herpesvirus 3, Elephantid herpesvirus 4, Elephantid herpesvirus 5, Procavid herpesvirus 1, Trichechid herpesvirus 1[4] and Common bottlenose dolphin gammaherpesvirus 1.[5]
Genera
Gammaherpesvirinae consists of the following seven genera:[2]
Additionally, the following three species are unassigned to a genus:[2]
Structure
Viruses in Gammaherpesvirinae are enveloped, with icosahedral, spherical to pleomorphic, and round geometries, and T=16 symmetry. The diameter is around 150-200 nm. Genomes are linear and non-segmented, around 180kb in length.[1]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Macavirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
| Percavirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
| Lymphocryptovirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
| Rhadinovirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
Life cycle
The main stages in the lifecycle of Gamma herpes virus are namely
• Virus attachment and entry
• Viral DNA injection through nuclear pore complex (NPC) into nucleus
• Assembly of nucleocapsids and encapsidation of viral genome
• Primary envelopment, invaginations of nuclear membranes and nuclear egress
• Tegumentation and secondary envelopment in the cytoplasm
• Egress and extracellular virions release
[6][7]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Macavirus | Mammals | T-lymphocytes | Glycoproteins | Budding | Nucleus | Nucleus | Nasal and ocular secretion |
| Percavirus | Mammals | B-lymphocytes | Glycoproteins | Budding | Nucleus | Nucleus | Sex; saliva |
| Lymphocryptovirus | Humans; mammals | B-lymphocytes | Glycoproteins | Budding | Nucleus | Nucleus | Saliva |
| Rhadinovirus | Humans; mammals | B-lymphocytes | Glycoproteins | Budding | Nucleus | Nucleus | Sex; saliva |
Lytic cycle
The lytic cycle of the gammaherpesviruses is initiated only on rare occasions.[7][8] Therefore, the least contribution to pathogenicity has to be expected from this stage. The ORFs expressed during that stage are further divided into immediate-early, early, and late. Promoter activation mediated by these proteins has also a strong effect on DNA synthesis from the origins of lytic DNA replication. As a result, virions are generated and released from the productively infected cells.[9]
Immune evasion strategies
Viruses that establish lifelong latent infections must ensure that the viral genome is maintained within the latently infected cell throughout the life of the host, yet at the same time must also be capable of avoiding elimination by the immune surveillance system especially must avoid being detected by host CD8+ cytotoxic T lymphocytes (CTLs). The gamma-herpesviruses are characteristically latent in lymphocytes and drive the proliferation that requires the expression of latent viral antigens.[10] The majority of gammaherpesviruses encode a specific protein that is critical for maintenance of the viral genome within latently infected cells termed the genome maintenance protein (GMP). During latency, the genome persists in the nucleus of the infected cells as a circular episomal element. GMPs are DNA-binding proteins that ensure that, as the host cell progresses through mitosis, the viral episomes are partitioned to daughter cells. This provides continuous existence of the viral genome within the host cells.[11][12]
MHV68 in genetically modified mice is used as a model for studying infection and host responses. Stable lifelong latency is a hallmark of the chronic phase of MHV68 infection. The spleen is a major infection site. The episomal maintenance protein for MHV68 is a latency-associated nuclear antigen (mLANA; ORF73). M2 protein mediates signaling pathways of infected B cells by interacting with SH2- and SH3-containing proteins. Host factors have been identified that can promote or antagonize MHV68 latency and reactivation.[7]
Human health
Gammaherpesviruses are of primary interest due to the two human viruses, EBV (Epstein–Barr virus) and KSHV (Kaposi's sarcoma-associated herpesvirus) and the diseases they cause. The gammaherpesviruses replicate and persist in lymphoid cells but some are capable of undergoing lytic replication in epithelial or fibroblast cells. Gammaherpesviruses may be a cause of chronic fibrotic lung diseases in humans and in animals.[13]
Murid herpesvirus 68 is an important model system for the study of gammaherpesviruses with tractable genetics.[7] The gammaherpesviruses, including HVS, EBV, KSHV, and RRV, are capable of establishing latent infection in lymphocytes.[11]
Attenuated virus mutants represent a promising approach towards gamma-herpesvirus infection control. Surprisingly, latency-deficient and, therefore, apathogenic MHV-68 mutants are found to be highly effective vaccines against these viruses.[10] Research in this area is almost exclusively performed using MHV68 as KSHV and EBV (the major human pathogens of this family) do not productively infect model organisms typically used for this type of experimentation.
Growth de-regulating genes
Herpesviruses have large genomes containing a wide array of genes. Although the first ORF in these gammaherpesviruses have oncogenic potential, other viral genes may also play a role in viral transformation. A striking feature of the four gammaherpesviruses is that they contain distinct ORFs involved in lymphocyte signaling events. At the left end of each viral genome are located ORFs encoding distinct transforming proteins. Gammaherpesvirus genes are capable of modulating cellular signals such that cell proliferation and viral replication occur at the appropriate times in the viral life cycle.[11]
References
- ↑ 1.0 1.1 "Viral Zone". ExPASy. http://viralzone.expasy.org/all_by_species/18.html.
- ↑ 2.0 2.1 2.2 "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. https://ictv.global/taxonomy.
- ↑ Nagamine B, Jones L, Tellgren-Roth C, Cavender J, Bratanich AC (2011) A novel gammaherpesvirus isolated from a black-tailed prairie dog (Cynomys ludovicianus). Arch Virol
- ↑ Wellehan JF, Johnson AJ, Childress AL, Harr KE, Isaza R (2008) Six novel gammaherpesviruses of Afrotheria provide insight into the early divergence of the Gammaherpesvirinae. Vet Microbiol 127(3-4):249-257
- ↑ Davison AJ, Subramaniam K, Kerr K, Jacob JM, Landrau-Giovannetti N, Walsh MT, Wells RS, Waltzek TB (2017) Genome sequence of a gammaherpesvirus from a common bottlenose dolphin (Tursiops truncatus). Genome Announc 5(31)
- ↑ Peng, L.; Ryazantsev, S.; Sun, R.; Zhou, Z. H. (2010). "Three-Dimensional Visualization of Gammaherpesvirus Life Cycle in Host Cells by Electron Tomography". Structure 18 (1): 47–58. doi:10.1016/j.str.2009.10.017. PMID 20152152.
- ↑ 7.0 7.1 7.2 7.3 Wang, Yiping; Tibbetts, Scott A.; Krug, Laurie T. (29 September 2021). "Conquering the Host: Determinants of Pathogenesis Learned from Murine Gammaherpesvirus 68". Annual Review of Virology 8 (1): 349–371. doi:10.1146/annurev-virology-011921-082615. ISSN 2327-056X. PMID 34586873.
- ↑ Oehmig, A.; Fraefel, C.; Breakefield, X. (2004). "Update on herpesvirus amplicon vectors". Mol Ther 10 (4): 630–643. doi:10.1016/j.ymthe.2004.06.641. PMID 15451447. https://cyberleninka.org/article/n/213700. Retrieved 10 November 2021.
- ↑ Ackermann, M. (2006). "Pathogenesis of gammaherpesvirus infections". Veterinary Microbiology 113 (3–4): 211–222. doi:10.1016/j.vetmic.2005.11.008. PMID 16332416.
- ↑ 10.0 10.1 Stevenson, P. G. (2004). "Immune evasion by gamma-herpesviruses". Current Opinion in Immunology 16 (4): 456–462. doi:10.1016/j.coi.2004.05.002. PMID 15245739.
- ↑ 11.0 11.1 11.2 Blake, N. (2010). "Immune evasion by gammaherpesvirus genome maintenance proteins". Journal of General Virology 91 (4): 829–846. doi:10.1099/vir.0.018242-0. PMID 20089802.
- ↑ Sorel, Océane; Dewals, Benjamin G. (2019). "The Critical Role of Genome Maintenance Proteins in Immune Evasion During Gammaherpesvirus Latency". Frontiers in Microbiology 9: 3315. doi:10.3389/fmicb.2018.03315. ISSN 1664-302X. PMID 30687291.
- ↑ Williams, KJ (March 2014). "Gammaherpesviruses and Pulmonary Fibrosis: Evidence From Humans, Horses, and Rodents". Veterinary Pathology 51 (2): 372–384. doi:10.1177/0300985814521838. PMID 24569614.
External links
- Gammaherpesvirinae at the US National Library of Medicine Medical Subject Headings (MeSH)
- ICTVdb Overview
- ICTVdb Details
- Taxonomic Proposals from the Herpesviridae Study Group
- Viralzone: Gammaherpesvirinae
- ICTV
Wikidata ☰ Q389837 entry
 |

