Biology:Protein kinase A
| cAMP-dependent protein kinase (Protein kinase A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
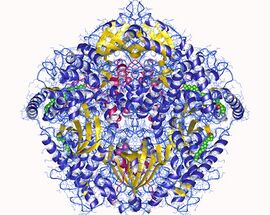 cAMP-dependent protein kinase hetero12mer, Sus scrofa | |||||||||
| Identifiers | |||||||||
| EC number | 2.7.11.11 | ||||||||
| CAS number | 142008-29-5 | ||||||||
| Alt. names | STK22, PKA, PKA C | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
In cell biology, protein kinase A (PKA) is a family of serine-threonine kinase[1] whose activity is dependent on cellular levels of cyclic AMP (cAMP). PKA is also known as cAMP-dependent protein kinase (EC 2.7.11.11). PKA has several functions in the cell, including regulation of glycogen, sugar, and lipid metabolism. It should not be confused with 5'-AMP-activated protein kinase (AMP-activated protein kinase).
History
Protein kinase A, more precisely known as adenosine 3',5'-monophosphate (cyclic AMP)-dependent protein kinase, abbreviated to PKA, was discovered by chemists Edmond H. Fischer and Edwin G. Krebs in 1968. They won the Nobel Prize in Physiology or Medicine in 1992 for their work on phosphorylation and dephosphorylation and how it relates to PKA activity.[2]
PKA is one of the most widely researched protein kinases, in part because of its uniqueness; out of 540 different protein kinase genes that make up the human kinome, only one other protein kinase, casein kinase 2, is known to exist in a physiological tetrameric complex, meaning it consists of four subunits.[1]
The diversity of mammalian PKA subunits was realized after Dr. Stan McKnight and others identified four possible catalytic subunit genes and four regulatory subunit genes. In 1991, Susan Taylor and colleagues crystallized the PKA Cα subunit, which revealed the bi-lobe structure of the protein kinase core for the very first time, providing a blueprint for all the other protein kinases in a genome (the kinome).[3]
Structure
When inactive, the PKA apoenzyme exists as a tetramer which consists of two regulatory subunits and two catalytic subunits. The catalytic subunit contains the active site, a series of canonical residues found in protein kinases that bind and hydrolyse ATP, and a domain to bind the regulatory subunit. The regulatory subunit has domains to bind to cyclic AMP, a domain that interacts with catalytic subunit, and an auto inhibitory domain. There are two major forms of regulatory subunit; RI and RII.[4]
Mammalian cells have at least two types of PKAs: type I is mainly in the cytosol, whereas type II is bound via its regulatory subunits and special anchoring proteins, described in the anchorage section, to the plasma membrane, nuclear membrane, mitochondrial outer membrane, and microtubules. In both types, once the catalytic subunits are freed and active, they can migrate into the nucleus (where they can phosphorylate transcription regulatory proteins), while the regulatory subunits remain in the cytoplasm.[5]
The following human genes encode PKA subunits:
- catalytic subunit – PRKACA, PRKACB, PRKACG
- regulatory subunit type I - PRKAR1A, PRKAR1B
- regulatory subunit type II - PRKAR2A, PRKAR2B
Mechanism
Activation
PKA is also commonly known as cAMP-dependent protein kinase, because it has traditionally been thought to be activated through release of the catalytic subunits when levels of the second messenger called cyclic adenosine monophosphate, or cAMP, rise in response to a variety of signals. However, recent studies evaluating the intact holoenzyme complexes, including regulatory AKAP-bound signalling complexes, have suggested that the local sub cellular activation of the catalytic activity of PKA might proceed without physical separation of the regulatory and catalytic components, especially at physiological concentrations of cAMP.[6][7] In contrast, experimentally induced supra physiological concentrations of cAMP, meaning higher than normally observed in cells, are able to cause separation of the holoenzymes, and release of the catalytic subunits.[6]
Extracellular hormones, such as glucagon and epinephrine, begin an intracellular signalling cascade that triggers protein kinase A activation by first binding to a G protein–coupled receptor (GPCR) on the target cell. When a GPCR is activated by its extracellular ligand, a conformational change is induced in the receptor that is transmitted to an attached intracellular heterotrimeric G protein complex by protein domain dynamics. The Gs alpha subunit of the stimulated G protein complex exchanges GDP for GTP in a reaction catalyzed by the GPCR and is released from the complex. The activated Gs alpha subunit binds to and activates an enzyme called adenylyl cyclase, which, in turn, catalyzes the conversion of ATP into cAMP, directly increasing the cAMP level. Four cAMP molecules are able to bind to the two regulatory subunits. This is done by two cAMP molecules binding to each of the two cAMP binding sites (CNB-B and CNB-A) which induces a conformational change in the regulatory subunits of PKA, causing the subunits to detach and unleash the two, now activated, catalytic subunits.[8]
Once released from inhibitory regulatory subunit, the catalytic subunits can go on to phosphorylate a number of other proteins in the minimal substrate context Arg-Arg-X-Ser/Thr.,[9] although they are still subject to other layers of regulation, including modulation by the heat stable pseudosubstrate inhibitor of PKA, termed PKI.[7][10]
Below is a list of the steps involved in PKA activation:
- Cytosolic cAMP increases
- Two cAMP molecules bind to each PKA regulatory subunit
- The regulatory subunits move out of the active sites of the catalytic subunits and the R2C2 complex dissociates
- The free catalytic subunits interact with proteins to phosphorylate Ser or Thr residues.
Catalysis
The liberated catalytic subunits can then catalyze the transfer of ATP terminal phosphates to protein substrates at serine, or threonine residues. This phosphorylation usually results in a change in activity of the substrate. Since PKAs are present in a variety of cells and act on different substrates, PKA regulation and cAMP regulation are involved in many different pathways.
The mechanisms of further effects may be divided into direct protein phosphorylation and protein synthesis:
- In direct protein phosphorylation, PKA directly either increases or decreases the activity of a protein.
- In protein synthesis, PKA first directly activates CREB, which binds the cAMP response element (CRE), altering the transcription and therefore the synthesis of the protein. In general, this mechanism takes more time (hours to days).
Phosphorylation mechanism
The Serine/Threonine residue of the substrate peptide is orientated in such a way that the hydroxyl group faces the gamma phosphate group of the bound ATP molecule. Both the substrate, ATP, and two Mg2+ ions form intensive contacts with the catalytic subunit of PKA. In the active conformation, the C helix packs against the N-terminal lobe and the Aspartate residue of the conserved DFG motif chelates the Mg2+ ions, assisting in positioning the ATP substrate. The triphosphate group of ATP points out of the adenosine pocket for the transfer of gamma-phosphate to the Serine/Threonine of the peptide substrate. There are several conserved residues, include Glutamate (E) 91 and Lysine (K) 72, that mediate the positioning of alpha- and beta-phosphate groups. The hydroxyl group of the peptide substrate's Serine/Threonine attacks the gamma phosphate group at the phosphorus via an SN2 nucleophilic reaction, which results in the transfer of the terminal phosphate to the peptide substrate and cleavage of the phosphodiester bond between the beta-phosphate and the gamma-phosphate groups. PKA acts as a model for understanding protein kinase biology, with the position of the conserved residues helping to distinguish the active protein kinase and inactive pseudokinase members of the human kinome.
Inactivation
Downregulation of protein kinase A occurs by a feedback mechanism and uses a number of cAMP hydrolyzing phosphodiesterase (PDE) enzymes, which belong to the substrates activated by PKA. Phosphodiesterase quickly converts cAMP to AMP, thus reducing the amount of cAMP that can activate protein kinase A. PKA is also regulated by a complex series of phosphorylation events, which can include modification by autophosphorylation and phosphorylation by regulatory kinases, such as PDK1.[7]
Thus, PKA is controlled, in part, by the levels of cAMP. Also, the catalytic subunit itself can be down-regulated by phosphorylation.
Anchorage
The regulatory subunit dimer of PKA is important for localizing the kinase inside the cell. The dimerization and docking (D/D) domain of the dimer binds to the A-kinase binding (AKB) domain of A-kinase anchor protein (AKAP). The AKAPs localize PKA to various locations (e.g., plasma membrane, mitochondria, etc.) within the cell.
AKAPs bind many other signaling proteins, creating a very efficient signaling hub at a certain location within the cell. For example, an AKAP located near the nucleus of a heart muscle cell would bind both PKA and phosphodiesterase (hydrolyzes cAMP), which allows the cell to limit the productivity of PKA, since the catalytic subunit is activated once cAMP binds to the regulatory subunits.
Function
PKA phosphorylates proteins that have the motif Arginine-Arginine-X-Serine exposed, in turn (de)activating the proteins. Many possible substrates of PKA exist; a list of such substrates is available and maintained by the NIH.[11]
As protein expression varies from cell type to cell type, the proteins that are available for phosphorylation will depend upon the cell in which PKA is present. Thus, the effects of PKA activation vary with cell type:
Overview table
| Cell type | Organ/system | Stimulators ligands → Gs-GPCRs or PDE inhibitors |
Inhibitors ligands → Gi-GPCRs or PDE stimulators |
Effects |
|---|---|---|---|---|
| adipocyte |
|
|||
| myocyte (skeletal muscle) | muscular system |
|
| |
| myocyte (cardiac muscle) | cardiovascular |
|
| |
| myocyte (smooth muscle) | cardiovascular |
|
|
Contributes to vasodilation (phosphorylates, and thereby inactivates, Myosin light-chain kinase) |
| hepatocyte | liver |
|
| |
| neurons in nucleus accumbens | nervous system | dopamine → dopamine receptor | Activate reward system | |
| principal cells in kidney | kidney |
|
| |
| Thick ascending limb cell | kidney | Vasopressin → V2 receptor | stimulate Na-K-2Cl symporter (perhaps only minor effect)[14] | |
| Cortical collecting tubule cell | kidney | Vasopressin → V2 receptor | stimulate Epithelial sodium channel (perhaps only minor effect)[14] | |
| Inner medullary collecting duct cell | kidney | Vasopressin → V2 receptor |
| |
| proximal convoluted tubule cell | kidney | PTH → PTH receptor 1 | Inhibit NHE3 → ↓H+ secretion[16] | |
| juxtaglomerular cell | kidney |
|
renin secretion |
In adipocytes and hepatocytes
Epinephrine and glucagon affect the activity of protein kinase A by changing the levels of cAMP in a cell via the G-protein mechanism, using adenylate cyclase. Protein kinase A acts to phosphorylate many enzymes important in metabolism. For example, protein kinase A phosphorylates acetyl-CoA carboxylase and pyruvate dehydrogenase. Such covalent modification has an inhibitory effect on these enzymes, thus inhibiting lipogenesis and promoting net gluconeogenesis. Insulin, on the other hand, decreases the level of phosphorylation of these enzymes, which instead promotes lipogenesis. Recall that gluconeogenesis does not occur in myocytes.
In nucleus accumbens neurons
PKA helps transfer/translate the dopamine signal into cells in the nucleus accumbens, which mediates reward, motivation, and task salience. The vast majority of reward perception involves neuronal activation in the nucleus accumbens, some examples of which include sex, recreational drugs, and food. Protein Kinase A signal transduction pathway helps in modulation of ethanol consumption and its sedative effects. A mouse study reports that mice with genetically reduced cAMP-PKA signalling results into less consumption of ethanol and are more sensitive to its sedative effects.[18]
In skeletal muscle
PKA is directed to specific sub-cellular locations after tethering to AKAPs. Ryanodine receptor (RyR) co-localizes with the muscle AKAP and RyR phosphorylation and efflux of Ca2+ is increased by localization of PKA at RyR by AKAPs.[19]
In cardiac muscle
In a cascade mediated by a GPCR known as β1 adrenoceptor, activated by catecholamines (notably norepinephrine), PKA gets activated and phosphorylates numerous targets, namely: L-type calcium channels, phospholamban, troponin I, myosin binding protein C, and potassium channels. This increases inotropy as well as lusitropy, increasing contraction force as well as enabling the muscles to relax faster.[20][21]
In memory formation
PKA has always been considered important in formation of a memory. In the fruit fly, reductions in expression activity of DCO (PKA catalytic subunit encoding gene) can cause severe learning disabilities, middle term memory and short term memory. Long term memory is dependent on the CREB transcription factor, regulated by PKA. A study done on drosophila reported that an increase in PKA activity can affect short term memory. However, a decrease in PKA activity by 24% inhibited learning abilities and a decrease by 16% affected both learning ability and memory retention. Formation of a normal memory is highly sensitive to PKA levels.[22]
See also
- Protein kinase
- Signal transduction
- G protein-coupled receptor
- Serine/threonine-specific protein kinase
- Myosin light-chain kinase
- cAMP-dependent pathway
References
- ↑ 1.0 1.1 Turnham, Rigney E.; Scott, John D. (2016-02-15). "Protein kinase A catalytic subunit isoform PRKACA; History, function and physiology". Gene 577 (2): 101–108. doi:10.1016/j.gene.2015.11.052. PMID 26687711.
- ↑ Knighton, D. R.; Zheng, J. H.; Ten Eyck, L. F.; Xuong, N. H.; Taylor, S. S.; Sowadski, J. M. (1991-07-26). "Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase". Science 253 (5018): 414–420. doi:10.1126/science.1862343. ISSN 0036-8075. PMID 1862343. Bibcode: 1991Sci...253..414K.
- ↑ Manning, G.; Whyte, D. B.; Martinez, R.; Hunter, T.; Sudarsanam, S. (2002-12-06). "The protein kinase complement of the human genome". Science 298 (5600): 1912–1934. doi:10.1126/science.1075762. ISSN 1095-9203. PMID 12471243. Bibcode: 2002Sci...298.1912M.
- ↑ "Kinase- and phosphatase-anchoring proteins: harnessing the dynamic duo" (in en). Nature Cell Biology 4 (8): E203–6. August 2002. doi:10.1038/ncb0802-e203. PMID 12149635.
- ↑ Alberts, Bruce (18 November 2014). Molecular biology of the cell (Sixth ed.). New York. p. 835. ISBN 978-0-8153-4432-2. OCLC 887605755.
- ↑ 6.0 6.1 Smith, FD; Esseltine, JL; Nygren, PJ; Veesler, D; Byrne, DP; Vonderach, M; Strashnov, I; Eyers, CE et al. (2017). "Local protein kinase A action proceeds through intact holoenzymes". Science 356 (6344): 1288–1293. doi:10.1126/science.aaj1669. PMID 28642438. PMC 5693252. Bibcode: 2017Sci...356.1288S. http://man.ac.uk/zcrk3W.
- ↑ 7.0 7.1 7.2 Byrne, DP; Vonderach, M; Ferries, S; Brownridge, PJ; Eyers, CE; Eyers, PA (2016). "cAMP-dependent protein kinase (PKA) complexes probed by complementary differential scanning fluorimetry and ion mobility-mass spectrometry". Biochemical Journal 473 (19): 3159–3175. doi:10.1042/bcj20160648. PMID 27444646.
- ↑ Lodish (2016). "15.5". Molecular Cell Biology (8th ed.). W.H. Freeman and Company. pp. 701. ISBN 978-1-4641-8339-3.
- ↑ Voet, Voet & Pratt (2008). Fundamentals of Biochemistry, 3rd Edition. Wiley. Pg 432
- ↑ Scott, JD; Glaccum, MB; Fischer, EH; Krebs, EG (1986). "Primary-structure requirements for inhibition by the heat-stable inhibitor of the cAMP-dependent protein kinase". PNAS 83 (6): 1613–1616. doi:10.1073/pnas.83.6.1613. PMID 3456605. Bibcode: 1986PNAS...83.1613S.
- ↑ "PKA Substrates". NIH. https://esbl.nhlbi.nih.gov/Databases/PKA-Targets/.
- ↑ 12.0 12.1 12.2 12.3 12.4 Pharmacology. Edinburgh: Churchill Livingstone. 2003. ISBN 978-0-443-07145-4. Page 172
- ↑ "Phospholamban: a key determinant of cardiac function and dysfunction". Archives des Maladies du Coeur et des Vaisseaux 98 (12): 1239–43. December 2005. PMID 16435604.
- ↑ 14.0 14.1 14.2 14.3 14.4 Boron, Walter F.; Boulpaep, Emile L. (2005). Medical Physiology: A Cellular And Molecular Approach (Updated ed.). Philadelphia, Pa.: Elsevier Saunders. p. 842. ISBN 978-1-4160-2328-9.
- ↑ Boron, Walter F.; Boulpaep, Emile L. (2005). Medical Physiology: A Cellular And Molecular Approaoch (Updated ed.). Philadelphia, Pa.: Elsevier Saunders. p. 844. ISBN 978-1-4160-2328-9.
- ↑ Boron, Walter F.; Boulpaep, Emile L. (2005). Medical Physiology: A Cellular And Molecular Approach (Updated ed.). Philadelphia, Pa.: Elsevier Saunders. p. 852. ISBN 978-1-4160-2328-9.
- ↑ 17.0 17.1 17.2 17.3 Boron, Walter F.; Boulpaep, Emile L. (2005). Medical Physiology: A Cellular And Molecular Approach (Updated ed.). Philadelphia, Pa.: Elsevier Saunders. p. 867. ISBN 978-1-4160-2328-9.
- ↑ Wand, Gary; Levine, Michael; Zweifel, Larry; Schwindinger, William; Abel, Ted (2001-07-15). "The cAMP–Protein Kinase A Signal Transduction Pathway Modulates Ethanol Consumption and Sedative Effects of Ethanol" (in en). Journal of Neuroscience 21 (14): 5297–5303. doi:10.1523/JNEUROSCI.21-14-05297.2001. ISSN 0270-6474. PMID 11438605.
- ↑ Ruehr, Mary L.; Russell, Mary A.; Ferguson, Donald G.; Bhat, Manju; Ma, Jianjie; Damron, Derek S.; Scott, John D.; Bond, Meredith (2003-07-04). "Targeting of Protein Kinase A by Muscle A Kinase-anchoring Protein (mAKAP) Regulates Phosphorylation and Function of the Skeletal Muscle Ryanodine Receptor" (in en). Journal of Biological Chemistry 278 (27): 24831–24836. doi:10.1074/jbc.M213279200. ISSN 0021-9258. PMID 12709444.
- ↑ Shah, Ajay M.; Solaro, R. John; Layland, Joanne (2005-04-01). "Regulation of cardiac contractile function by troponin I phosphorylation" (in en). Cardiovascular Research 66 (1): 12–21. doi:10.1016/j.cardiores.2004.12.022. ISSN 0008-6363. PMID 15769444.
- ↑ Boron, Walter F.; Boulpaep, Emile L. (2012). Medical physiology : a cellular and molecular approach. Boron, Walter F.,, Boulpaep, Emile L. (Updated second ed.). Philadelphia, PA. ISBN 9781437717532. OCLC 756281854.
- ↑ Horiuchi, Junjiro; Yamazaki, Daisuke; Naganos, Shintaro; Aigaki, Toshiro; Saitoe, Minoru (2008-12-30). "Protein kinase A inhibits a consolidated form of memory in Drosophila" (in en). Proceedings of the National Academy of Sciences 105 (52): 20976–20981. doi:10.1073/pnas.0810119105. ISSN 0027-8424. PMID 19075226. Bibcode: 2008PNAS..10520976H.
External links
- Cyclic+AMP-Dependent+Protein+Kinases at the US National Library of Medicine Medical Subject Headings (MeSH)
- Drosophila cAMP-dependent protein kinase 1 - The Interactive Fly
- cAMP-dependent protein kinase: PDB Molecule of the Month
- Overview of all the structural information available in the PDB for UniProt: P25321 (cAMP-dependent protein kinase catalytic subunit alpha) at the PDBe-KB.
Notes
 |