Biology:Potyvirus
| Potyvirus | |
|---|---|
| |
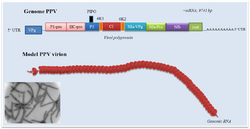
| Plum pox virus genome with electron micrograph and model of virions | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Stelpaviricetes |
| Order: | Patatavirales |
| Family: | Potyviridae |
| Genus: | Potyvirus |
| Species | |
|
See text | |
Potyvirus is a genus of positive-strand RNA viruses (named after its type species, Potato virus Y (PVY)) in the family Potyviridae. Plants serve as natural hosts. Like begomoviruses, members of this genus may cause significant losses in agricultural, pastoral, horticultural, and ornamental crops. More than 200 species of aphids spread potyviruses, and most are from the subfamily Aphidinae (genera Macrosiphum and Myzus).[citation needed] The genus contains 190 species and potyviruses account for about thirty percent of all currently known plant viruses.[1][2]
Structure
The virion is non-enveloped with a flexuous and filamentous nucleocapsid, 680 to 900 nanometers (nm) long and is 11–20 nm in diameter.[1] The nucleocapsid contains around 2000 copies of the capsid protein. The symmetry of the nucleocapsid is helical with a pitch of 3.4-3.5 nm.[1]
Genome
The genome is a linear, positive-sense, single-stranded RNA ranging in size from 9,000 to 12,000 nucleotide bases. Most potyviruses have non-segmented genomes,[1] though a number of species are bipartite. The typical base compositions of some of the most common, non-recombinant strains of the type species, PVY, range between ~23.4-23.8 % G; ~31-31.6 % A; ~18.2-18.8 % C; and ~26.5-26.8 % U.[3]
In the species with a monopartite genome, a genome-linked VPg protein is covalently bound to the 5' end and the 3' end is polyadenylated. The genome encodes a single open reading frame (ORF) expressed as a 350 kDa polyprotein precursor. This polyprotein is processed into ten smaller proteins: protein 1 protease (P1-Pro), helper component protease (HC-Pro), protein 3 (P3), cylindrical inclusion (CI), viral protein genome-linked (Vpg), nuclear inclusion A (NIa), nuclear inclusion B (NIb), capsid protein (CP) and two small putative proteins known as 6K1 and 6K2. The P3 cistron also contains an overlapping reading frame called "Pretty interesting Potyviridae ORF" (PIPO).[4] PIPO codes for an alternative C-terminus to the P3 protein, which is generated into a subset of transcripts by a +2 frameshift caused by a ribosome slippage mechanism at a conserved GA6 repeat sequence.[5][6] The resulting protein is called P3N-PIPO. A similar mechanism is thought to produce an alternative reading frame within the P1 cistron, named "pretty interesting sweet potato potyvirus ORF" (PISPO), in a number of sweet potato-infecting potyviruses including sweet potato feathery mottle virus.[7]
Proteome
P1 (~33 kilodaltons (kDa) in molecular weight) is a serine protease which facilitates its own cleavage from the polyprotein at the P1-HC-Pro junction.[8] P1 consists of a conserved C-terminal protease domain and an N-terminal region which has a high level of variation in sequence and length between potyvirus species but exhibits conserved patterns of intrinsic disorder. P1 is also promotes viral RNA replication, though it is not required for it.[9]
HC-Pro (~52 KDa) is a cysteine protease which cleaves a glycine-glycine dipeptide at its own C-terminus.[8] It also interacts with eukaryotic initiation factor 4 (eIF4). It acts as a viral RNA silencing suppressor through its interactions with host AGO proteins.[10] HC-Pro's activity is regulated by the adjacent P1 protein: before P1 cleaves itself off the P1-HC-Pro intermediate, the P1 terminus reduces HC-Pro's RNA silencing suppression activity.[8] The rate of P1 cleavage therefore regulates the level of RNA interference suppression during infection. HC-Pro is also involved in aphid transmission.[11] Though the exact mechanism is unknown, HC-Pro has been proposed to attach to host aphid mouth parts through its N-terminal zinc finger-like domain and anchor virions through its interactions with the capsid protein.[12]
P3 (~41 kDa) is a membrane protein which is required for viral replication and accumulates in viral replication vesicles.[13] It mediates the interactions between replication vesicles and movement complex proteins which may allow replication vesicles to be recruited to the movement complex for efficient intercellular movement.[14] P3 also interacts with large subunit of the ribulose-1,5-bisphosphate carboxylase/oxygenase.[citation needed]
CI (~71 kDa) is an RNA helicase with ATPase activity.[15] Its most unusual property is its ability to form large and highly symmetrical conical and cylindrical inclusions with a central hollow cylinder from which laminate sheets radiate outward and fold in on themselves in a pattern often described as "pinwheels". These inclusions are easily seen in transmission electron micrographs of infected tissues and were historically used as a diagnostic criterion for potyvirus infections. CI inclusions are a major component of the potyviral movement complex which is assembled at plasmodesmata. CI is also required for viral replication and is present on replication membranes. Its exact contributions to replication are not clear but, as an RNA helicase, CI is likely facilitating replication by dismantling the secondary structures of viral RNA.
NIa (~50 kDa) forms crystalline inclusions in the host nucleus. It is cleaved into NIa-Pro and VPg.
NIa-Pro (~27 kDa) is a cysteine protease which processes most of the cleavage sites of the polyprotein.[16] The only exceptions are the self-cleavages of P1 and HC-Pro. The high degree of cleavage sequence specificity and conservation has made NIa-Pro (often that of Tobacco etch virus) a valuable tool in biotechnology, especially in applications which require removing affinity tags from recombinant proteins after affinity purification. NIa-Pro has also shown to exhibit sequence-independent DNase activity and to interfere with host DNA methylation suggesting that NIa and/or NIa-Pro are altering in host gene expression.[17] Potyviral NIa-Pro shares a high level of homology with the picornaviral 3C protease.[18]
VPg (~22 kDa) is covalently attched to the 5' end of the viral genomic RNA through uridylation and is thought to act as a primer for viral genome replication similarly to the VPg proteins of picornaviridae.[19] It is a highly disordered protein and its flexibility has been suggested to allow it to interact with many other viral proteins. VPg also interacts with various host proteins including eukaryotic initiation factor 4E (eIF4E), eukaryotic elongation factor 1A (eEF1A), and poly(A)-binding protein (PABP).[20][21]
NIb (~59 kDa) is a superfamily II RNA-dependent RNA polymerase (RdRp) which polymerises viral RNA during replication.[22] Like NIa, NIb forms inclusions in the host nucleus where it is transported due to its two nuclear localisation sequences. NIb has the three-domain "palm, thumb, and fingers" structure typical of RdRps.
6K1 (~6 kDa) the function is not known, but because it accumulates in replication vesicles and has a transmembrane domain, 6K1 is thought to contribute to virus-induced vesicle formation.[23]
6K2 (~6 kDa) is a transmembrane protein which rearranges host membranes into virus-induced membrane structures.[24] It interacts with various ER exit site proteins to produce vesicular and tubular extensions which eventually mature into replication vesicles.[25] 6K2 has three main domains: the N-terminal domain which is required for cell-to-cell movement, the central hydrophobic transmembrane alpha helix, and the C terminal domain which is required for viral replication.[26]
P3N-PIPO (~25 kDa) is a dedicated movement protein which anchors the movement complex to the plasmodesma.[27] It may also modulate the plasmodesmatal size exclusion limit by interacting with host proteins which sever plasmodesmatal actin filaments and reduce callose deposition.[28][29] It interacts with both the large and small subunits of the ribulose-1,5-bisphosphate carboxylase/oxygenase.[citation needed]
CP (~30 - 35 kDa) is the capsid protein. It has two terminal domains which are disordered and exposed at the surface of the virion.[30][31] The central core domain contains an RNA-binding pocket which binds to viral RNA. The structure of the capsid protein is highly conserved in potyviruses, though there is a relatively high degree of sequence variability. In addition to encapsidating the virion, CP core domain is required for intercellular movement and contributes to seed transmission.[32]
Certain atypical potyviruses code for additional proteins or protein domains, such as P1-PISPO, Alkylation B (AlkB), and inosine triphosphate pyrophosphatase (known as ITPase or HAM1).[33] Such anomalies are often situated in the hypervariable P1-HC-Pro region.[8]
Life cycle
Transmission
Most potyviruses are transmitted by aphids as they probe plant tissues with their stylet during feeding.[34] They do not circulate or multiply within the aphid and typically only persist in the aphid for a few minutes. Certain potyviruses have been shown to alter the feeding patterns of their aphid vectors, which may manifest as longer periods of time spent on infected plants, reduced non-probing feeding time, and increased phloem sap ingestion.
Seed and pollen transmission has been documented in certain potyvirus species, for instance in PVY and Turnip mosaic virus (TUMV).[35] Vegetative transmission by infected tubers or grafting material are of particular concern for certain agricultural crops, such as potato and fruit trees, respectively.
Transmission can also occur by physical contact with infected plants or with contaminated tools, clothes, or even water.[36]
Translation
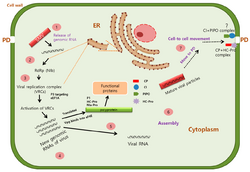
After entry, potyvirus particles are uncoated and genomic RNA is released into host cytoplasm. Potyviral RNA mimicks host mRNA: the 5' VPg protein shares functional similarities with the 5' cap and the 3' end is polyadenylated.[37] VPg and its interactions with eIF4E and eIF4(iso)E allow the virus to utilise host cap-dependent translation machinery for its translation. Similarly to eukaryotic translation, the VPg-eIF4E interaction assembles the eIF4F complex around viral RNA.
A number of weak internal ribosome entry sites (IRES) have been identified in many potyvirus species but it is not known whether cap-independent translation is an important translation mechanism for potyviruses.[37]
Replication
Like many other positive strand RNA viruses, potyvirus replication is heavily associated with host membranes.[1][38] The viral 6K2 protein coordinates the rearrangement of host membranes into various infection-associated structures which, depending of the potyvirus species, can include anything from small round viral vesicles to complex globular structures with many cisternae or lobes. These structures are dotted with viral replication complexes and are often called "replication vesicles", "viroplasm" or "viral factories". Replication vesicle membranes are derived from a variety of host organelles and the sources differ between potyvirus species. Some membrane sources include the ER, chloroplasts, Golgi apparatus, and vacuoles.
The exact replication mechanism is not known but it involves a negative sense RNA intermediate and requires both viral and host proteins. Viral proteins detected in replication complexes include HC-Pro, P3, 6K1, 6K2, CI, VPg, NIa-Pro, and NIb.[39] Host factors present in replication vesicles include eIF4A and several heat shock proteins.
Intercellular movement
Like most plant viruses, potyviruses have evolved to move from one plant cell to another through plasmodesmata. However, unlike some well-studied plant viruses, such as the Tobacco mosaic virus, potyviruses do not have a single movement protein but instead assemble a movement complex around the plasmodesma.[40] This complex is primarily composed of three viral proteins: CI, CP, P3N-PIPO. Conical CI inclusions are anchored to plasmodesmata by P3N-PIPO during the early stages of potyvirus infection. This allows the inclusion to funnel either viral particles or viral RNA-CP complexes through the plasmodesma. Replication vesicles are also recruited to the movement complex, suggesting that replication and movement are coupled. Replication vesicles are recruited by P3N-PIPO, which interacts with both CI and P3 through the shared P3N-domain.[14] P3's interaction with 6K2 allows replication vesicles to be tethered to the movement complex.
Evolution
Potyviruses evolved between 6,600 and 7,250 years ago.[41][42] They appear to have evolved in southwest Eurasia or north Africa. The estimated mutation rate is about 1.15×10−4 nucleotide substitutions/site/year.[citation needed]
Geographical distribution
Agriculture was introduced into Australia in the 18th century. This introduction also included plant pathogens. Thirty eight potyvirus species have been isolated in Australia. Eighteen potyviruses have been found only in Australia and are presumed to be endemic there. The remaining twenty appear to have been introduced with agriculture.[citation needed]
Diagnostics
Historically, potyvirus diagnostics relied on the detection of various proteinaceous inclusions in infected plant cells. These may appear as crystals in either the cytoplasm or in the nucleus, as amorphous X-bodies, membranous bodies, viroplasms or pinwheels.[43] The inclusions may or may not (depending on the species) contain virions.[citation needed] These inclusions can be seen by light microscopy in leaf strips of infected plant tissue stained with Orange-Green (protein stain) but not Azure A (nucleic acid stain).[44][45][46]
Modern detection methods rely primarily on reverse transcription PCR.[47]
Taxonomy
Potyvirus contains the following species:[2]
- African eggplant mosaic virus
- Algerian watermelon mosaic virus
- Alstroemeria mosaic virus
- Alternanthera mild mosaic virus
- Amaranthus leaf mottle virus
- Amazon lily mosaic virus
- Angelica virus Y
- Apium virus Y
- Araujia mosaic virus
- Arracacha mottle virus
- Asparagus virus 1
- Banana bract mosaic virus
- Barbacena virus Y
- Basella rugose mosaic virus
- Bean common mosaic necrosis virus
- Bean common mosaic virus
- Bean yellow mosaic virus
- Beet mosaic virus
- Begonia flower breaking virus
- Bidens mosaic virus
- Bidens mottle virus
- Blue squill virus A
- Brugmansia mosaic virus
- Brugmansia suaveolens mottle virus
- Butterfly flower mosaic virus
- Calanthe mild mosaic virus
- Calla lily latent virus
- Callistephus mottle virus
- Canna yellow streak virus
- Carnation vein mottle virus
- Carrot thin leaf virus
- Carrot virus Y
- Catharanthus mosaic virus
- Celery mosaic virus
- Ceratobium mosaic virus
- Chilli ringspot virus
- Chilli veinal mottle virus
- Chinese artichoke mosaic virus
- Clitoria virus Y
- Clover yellow vein virus
- Cocksfoot streak virus
- Colombian datura virus
- Commelina mosaic virus
- Costus stripe mosaic virus
- Cowpea aphid-borne mosaic virus
- Cucurbit vein banding virus
- Cypripedium virus Y
- Cyrtanthus elatus virus A
- Daphne mosaic virus
- Daphne virus Y
- Dasheen mosaic virus
- Datura shoestring virus
- Dendrobium chlorotic mosaic virus
- Dioscorea mosaic virus
- Diuris virus Y
- Donkey orchid virus A
- East Asian Passiflora distortion virus
- East Asian Passiflora virus
- Endive necrotic mosaic virus
- Euphorbia ringspot virus
- Freesia mosaic virus
- Fritillary virus Y
- Gloriosa stripe mosaic virus
- Gomphocarpus mosaic virus
- Habenaria mosaic virus
- Hardenbergia mosaic virus
- Henbane mosaic virus
- Hibbertia virus Y
- Hippeastrum mosaic virus
- Hyacinth mosaic virus
- Impatiens flower break virus
- Iris fulva mosaic virus
- Iris mild mosaic virus
- Iris severe mosaic virus
- Japanese yam mosaic virus
- Jasmine virus T
- Johnsongrass mosaic virus
- Kalanchoe mosaic virus
- Keunjorong mosaic virus
- Konjac mosaic virus
- Leek yellow stripe virus
- Lettuce Italian necrotic virus
- Lettuce mosaic virus
- Lily mottle virus
- Lily virus Y
- Lupinus mosaic virus
- Lycoris mild mottle virus
- Maize dwarf mosaic virus
- Malva vein clearing virus
- Mashua virus Y
- Meadow saffron breaking virus
- Mediterranean ruda virus
- Moroccan watermelon mosaic virus
- Narcissus degeneration virus
- Narcissus late season yellows virus
- Narcissus yellow stripe virus
- Nerine yellow stripe virus
- Noni mosaic virus
- Nothoscordum mosaic virus
- Onion yellow dwarf virus
- Ornithogalum mosaic virus
- Ornithogalum virus 2
- Ornithogalum virus 3
- Panax virus Y
- Papaya leaf distortion mosaic virus
- Papaya ringspot virus
- Paris mosaic necrosis virus
- Paris virus 1
- Parsnip mosaic virus
- Passiflora chlorosis virus
- Passiflora mottle virus
- Passion fruit woodiness virus
- Pea seed-borne mosaic virus
- Peanut mottle virus
- Pecan mosaic-associated virus
- Pennisetum mosaic virus
- Pepper mottle virus
- Pepper severe mosaic virus
- Pepper veinal mottle virus
- Pepper yellow mosaic virus
- Peru tomato mosaic virus
- Pfaffia mosaic virus
- Platycodon mild mottle virus
- Pleione flower breaking virus
- Pleione virus Y
- Plum pox virus
- Pokeweed mosaic virus
- Potato virus A
- Potato virus V
- Potato virus Y
- Potato yellow blotch virus
- Ranunculus leaf distortion virus
- Ranunculus mild mosaic virus
- Ranunculus mosaic virus
- Rhopalanthe virus Y
- Saffron latent virus
- Sarcochilus virus Y
- Scallion mosaic virus
- Shallot yellow stripe virus
- Sorghum mosaic virus
- Soybean mosaic virus
- Spiranthes mosaic virus 3
- Sudan watermelon mosaic virus
- Sugarcane mosaic virus
- Sunflower chlorotic mottle virus
- Sunflower mild mosaic virus
- Sunflower mosaic virus
- Sunflower ring blotch virus
- Sweet potato feathery mottle virus
- Sweet potato latent virus
- Sweet potato mild speckling virus
- Sweet potato virus 2
- Sweet potato virus C
- Sweet potato virus G
- Tamarillo leaf malformation virus
- Telfairia mosaic virus
- Telosma mosaic virus
- Thunberg fritillary mosaic virus
- Tobacco etch virus
- Tobacco mosqueado virus
- Tobacco vein banding mosaic virus
- Tobacco vein mottling virus
- Tomato necrotic stunt virus
- Tradescantia mild mosaic virus
- Tuberose mild mosaic virus
- Tuberose mild mottle virus
- Tulip breaking virus
- Tulip mosaic virus
- Turnip mosaic virus
- Twisted-stalk chlorotic streak virus
- Ugandan passiflora virus
- Vallota mosaic virus
- Vanilla distortion mosaic virus
- Verbena virus Y
- Watermelon leaf mottle virus
- Watermelon mosaic virus
- Wild melon banding virus
- Wild onion symptomless virus
- Wild potato mosaic virus
- Wild tomato mosaic virus
- Wisteria vein mosaic virus
- Yam mild mosaic virus
- Yam mosaic virus
- Yambean mosaic virus
- Zantedeschia mild mosaic virus
- Zea mosaic virus
- Zucchini shoestring virus
- Zucchini tigre mosaic virus
- Zucchini yellow fleck virus
- Zucchini yellow mosaic virus
A further four viruses were previously classified as species in this genus but were abolished due to lack of genetic sequence information:[48]
- Cowpea green vein banding virus
- Groundnut eyespot virus
- Helenium virus Y
- Tropaeolum mosaic virus
Species groups
Potyviruses were further divided into the PVY, SCMV, BYMV, BCMV species groups in 1992. Gibbs and Ohshima 2010 produced a more extensive molecular phylogeny with the same four, but also several new groups: the BtMV, ChVMV, DaMV, OYDV, PRSV, TEV, and TuMV.[42]
PVY
Contains 16 species including the type species of the genus (potato virus Y). The primary hosts are: Nine Solanaceae, three Amaranthus, three Asteraceae, one Lilium, and one Amaryllis.[42]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "ICTV Virus Taxonomy Profile: Potyviridae 2022". The Journal of General Virology 103 (5): 001738. May 2022. doi:10.1099/jgv.0.001738. PMID 35506996. https://researchrepository.murdoch.edu.au/id/eprint/64784/.
- ↑ 2.0 2.1 "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. https://ictv.global/taxonomy.
- ↑ "Analysis of codon usage bias in potato virus Y non-recombinant strains". Virus Research 286: 198077. September 2020. doi:10.1016/j.virusres.2020.198077. PMID 32619560.
- ↑ "An overlapping essential gene in the Potyviridae". Proceedings of the National Academy of Sciences of the United States of America 105 (15): 5897–5902. April 2008. doi:10.1073/pnas.0800468105. PMID 18408156. Bibcode: 2008PNAS..105.5897C.
- ↑ "RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the potyviridae family". Journal of Virology 89 (13): 6965–6967. July 2015. doi:10.1128/JVI.00337-15. PMID 25878117.
- ↑ "Transcriptional slippage in the positive-sense RNA virus family Potyviridae". EMBO Reports 16 (8): 995–1004. August 2015. doi:10.15252/embr.201540509. PMID 26113364.
- ↑ "A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing". Molecular Plant Pathology 17 (7): 1111–1123. September 2016. doi:10.1111/mpp.12366. PMID 26757490.
- ↑ 8.0 8.1 8.2 8.3 "The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses". PLOS Pathogens 10 (3): e1003985. March 2014. doi:10.1371/journal.ppat.1003985. PMID 24603811.
- ↑ "Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification". Journal of Virology 69 (6): 3668–3674. June 1995. doi:10.1128/jvi.69.6.3668-3674.1995. PMID 7745715.
- ↑ "The potyviral silencing suppressor HCPro recruits and employs host ARGONAUTE1 in pro-viral functions". PLOS Pathogens 16 (10): e1008965. October 2020. doi:10.1371/journal.ppat.1008965. PMID 33031436.
- ↑ "Helper-dependent vector transmission of plant viruses". Annual Review of Phytopathology 34 (1): 227–247. September 1996. doi:10.1146/annurev.phyto.34.1.227. PMID 15012542.
- ↑ "The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have". Molecular Plant Pathology 19 (3): 744–763. March 2018. doi:10.1111/mpp.12553. PMID 28371183.
- ↑ "The C-terminal region of the Turnip mosaic virus P3 protein is essential for viral infection via targeting P3 to the viral replication complex". Virology 510: 147–155. October 2017. doi:10.1016/j.virol.2017.07.016. PMID 28735115.
- ↑ 14.0 14.1 "P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain To Recruit Viral Replication Vesicles for Cell-to-Cell Movement". Journal of Virology 94 (8). March 2020. doi:10.1128/JVI.01898-19. PMID 31969439.
- ↑ "The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein". Molecular Plant-Microbe Interactions 27 (3): 215–226. March 2014. doi:10.1094/MPMI-11-13-0333-CR. PMID 24405034.
- ↑ "Expanding Repertoire of Plant Positive-Strand RNA Virus Proteases". Viruses 11 (1): 66. January 2019. doi:10.3390/v11010066. PMID 30650571.
- ↑ "The NIa-Protease Protein Encoded by the Pepper Mottle Virus Is a Pathogenicity Determinant and Releases DNA Methylation of Nicotiana benthamiana". Frontiers in Microbiology 11: 102. 2020-02-21. doi:10.3389/fmicb.2020.00102. PMID 32153517.
- ↑ "The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups". Nature Reviews. Microbiology 6 (12): 925–939. December 2008. doi:10.1038/nrmicro2030. PMID 18997823.
- ↑ "Hydrodynamic Behavior of the Intrinsically Disordered Potyvirus Protein VPg, of the Translation Initiation Factor eIF4E and of their Binary Complex". International Journal of Molecular Sciences 20 (7): 1794. April 2019. doi:10.3390/ijms20071794. PMID 30978975.
- ↑ "Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity". Journal of Virology 74 (17): 7730–7737. September 2000. doi:10.1128/jvi.74.17.7730-7737.2000. PMID 10933678.
- ↑ "Dissecting the molecular network of virus-plant interactions: the complex roles of host factors". Annual Review of Phytopathology 53 (1): 45–66. 2015-08-04. doi:10.1146/annurev-phyto-080614-120001. PMID 25938276.
- ↑ "The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection". Viruses 12 (1): 77. January 2020. doi:10.3390/v12010077. PMID 31936267.
- ↑ "Plum Pox Virus 6K1 Protein Is Required for Viral Replication and Targets the Viral Replication Complex at the Early Stage of Infection". Journal of Virology 90 (10): 5119–5131. May 2016. doi:10.1128/JVI.00024-16. PMID 26962227.
- ↑ "Cellular remodeling during plant virus infection". Annual Review of Phytopathology 48 (1): 69–91. 2010-07-01. doi:10.1146/annurev-phyto-073009-114239. PMID 20337516.
- ↑ "Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner". Journal of Virology 82 (24): 12252–12264. December 2008. doi:10.1128/JVI.01329-08. PMID 18842721.
- ↑ "Mutagenesis Scanning Uncovers Evolutionary Constraints on Tobacco Etch Potyvirus Membrane-Associated 6K2 Protein". Genome Biology and Evolution 11 (4): 1207–1222. April 2019. doi:10.1093/gbe/evz069. PMID 30918938.
- ↑ "Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO". PLOS Pathogens 6 (6): e1000962. June 2010. doi:10.1371/journal.ppat.1000962. PMID 20585568.
- ↑ "Remorin interacting with PCaP1 impairs Turnip mosaic virus intercellular movement but is antagonised by VPg". The New Phytologist 225 (5): 2122–2139. March 2020. doi:10.1111/nph.16285. PMID 31657467.
- ↑ "StREM1.3 REMORIN Protein Plays an Agonistic Role in Potyvirus Cell-to-Cell Movement in N. benthamiana". Viruses 14 (3): 574. March 2022. doi:10.3390/v14030574. PMID 35336981.
- ↑ "Structure of Turnip mosaic virus and its viral-like particles". Scientific Reports 9 (1): 15396. October 2019. doi:10.1038/s41598-019-51823-4. PMID 31659175. Bibcode: 2019NatSR...915396C.
- ↑ "Structural basis for the multitasking nature of the potato virus Y coat protein". Science Advances 5 (7): eaaw3808. July 2019. doi:10.1126/sciadv.aaw3808. PMID 31328164. Bibcode: 2019SciA....5.3808K.
- ↑ "Potyviral coat protein and genomic RNA: A striking partnership leading virion assembly and more". Virus Assembly and Exit Pathways. 108. Academic Press. January 2020. pp. 165–211. doi:10.1016/bs.aivir.2020.09.001. ISBN 9780128207611.
- ↑ "Proteome expansion in the Potyviridae evolutionary radiation". FEMS Microbiology Reviews 46 (4): fuac011. July 2022. doi:10.1093/femsre/fuac011. PMID 35195244.
- ↑ Gadhave, K R; Gautam, S; Rasmussen, D A; Srinivasan, R (2020-07-17). "Aphid Transmission of Potyvirus: The Largest Plant-Infecting RNA Virus Genus" (in en). Viruses 12 (7): 773. doi:10.3390/v12070773. ISSN 1999-4915. PMID 32708998.
- ↑ Simmons, H E; Munkvold, G P (2014), Gullino, M L; Munkvold, Gary, eds., "Seed Transmission in the Potyviridae" (in en), Global Perspectives on the Health of Seeds and Plant Propagation Material (Dordrecht: Springer Netherlands): pp. 3–15, doi:10.1007/978-94-017-9389-6_1, ISBN 978-94-017-9388-9, https://link.springer.com/10.1007/978-94-017-9389-6_1, retrieved 2023-08-16
- ↑ Mehle, N.; Gutiérrez-Aguirre, I.; Prezelj, N.; Delić, D.; Vidic, U.; Ravnikar, M. (2014-02-15). "Survival and Transmission of Potato Virus Y, Pepino Mosaic Virus, and Potato Spindle Tuber Viroid in Water" (in en). Applied and Environmental Microbiology 80 (4): 1455–1462. doi:10.1128/AEM.03349-13. ISSN 0099-2240. PMID 24334672. Bibcode: 2014ApEnM..80.1455M.
- ↑ 37.0 37.1 Jaramillo-Mesa, Helena; Rakotondrafara, Aurélie M. (2023-10-01). "All eggs in one basket: How potyvirus infection is controlled at a single cap-independent translation event". Seminars in Cell & Developmental Biology. Special Issue: Plant pathogens and disease susceptibility 148-149: 51–61. doi:10.1016/j.semcdb.2022.12.011. ISSN 1084-9521. PMID 36608998. https://www.sciencedirect.com/science/article/pii/S1084952122003792.
- ↑ Wei, Taiyun; Huang, Tyng-Shyan; McNeil, Jamie; Laliberté, Jean-François; Hong, Jian; Nelson, Richard S.; Wang, Aiming (2010-01-15). "Sequential Recruitment of the Endoplasmic Reticulum and Chloroplasts for Plant Potyvirus Replication" (in en). Journal of Virology 84 (2): 799–809. doi:10.1128/JVI.01824-09. ISSN 0022-538X. PMID 19906931.
- ↑ Lõhmus, Andres; Varjosalo, Markku; Mäkinen, Kristiina (August 2016). "Protein composition of 6K2-induced membrane structures formed during Potato virus A infection: PVA replication complex proteome" (in en). Molecular Plant Pathology 17 (6): 943–958. doi:10.1111/mpp.12341. PMID 26574906.
- ↑ Wang, Aiming (June 2021). "Cell-to-cell movement of plant viruses via plasmodesmata: a current perspective on potyviruses" (in en). Current Opinion in Virology 48: 10–16. doi:10.1016/j.coviro.2021.03.002. PMID 33784579. https://linkinghub.elsevier.com/retrieve/pii/S1879625721000183.
- ↑ "The prehistory of potyviruses: their initial radiation was during the dawn of agriculture". PLOS ONE 3 (6): e2523. June 2008. doi:10.1371/journal.pone.0002523. PMID 18575612. Bibcode: 2008PLoSO...3.2523G.
- ↑ 42.0 42.1 42.2 "Potyviruses and the digital revolution". Annual Review of Phytopathology (Annual Reviews) 48 (1): 205–223. 2010. doi:10.1146/annurev-phyto-073009-114404. PMID 20438367.
- ↑ Florida Department of Agriculture and Consumer Services: Florida plant viruses and their inclusions—Potyvirus
- ↑ "Materials and Methods for the Detection of Viral Inclusions". University of Florida - Institute of Food and Agricultural Sciences. http://www.doacs.state.fl.us/pi/enpp/pathology/florida_viruses/Howto.html.
- ↑ Christie, R.G. and Edwardson, J.R. (1977). Fla Agric. Exp. Stn Monog. No. 9, 150 pp.
- ↑ How do you diagnose a virus infection in a plant?
- ↑ Thomson, Darelle; Dietzgen, Ralf G. (August 1995). "Detection of DNA and RNA plant viruses by PCR and RT-PCR using a rapid virus release protocol without tissue homogenization" (in en). Journal of Virological Methods 54 (2–3): 85–95. doi:10.1016/0166-0934(95)00022-M. PMID 8530569. https://linkinghub.elsevier.com/retrieve/pii/016609349500022M.
- ↑ "Create three species in genus Potyvirus and abolish five species in genus Potyvirus". 2016. https://ictv.global/ictv/proposals/2016.008a,bP.A.v3.Potyvirus_3sprem.pdf.
Bibliography
- "Taxonomy of potyviruses: current problems and some solutions". Intervirology 32 (5): 269–296. 1991. doi:10.1159/000150211. PMID 1657820.
- "Potyvirus". Virus taxonomy : classification and nomenclature of viruses : ninth report of the International Committee on Taxonomy of Viruses. London: Academic Press. 2012. pp. 926–1072. ISBN 978-0123846846. https://books.google.com/books?id=KXRCYay3pH4C&pg=PA1072. Retrieved 9 December 2014.
External links
Wikidata ☰ Q3400142 entry
 |