Biology:Reverse transcription polymerase chain reaction
Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA (in this context called complementary DNA or cDNA) and amplification of specific DNA targets using polymerase chain reaction (PCR).[1] It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Confusion can arise because some authors use the acronym RT-PCR to denote real-time PCR. In this article, RT-PCR will denote Reverse Transcription PCR. Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.
The close association between RT-PCR and qPCR has led to metonymic use of the term qPCR to mean RT-PCR. Such use may be confusing,[2] as RT-PCR can be used without qPCR, for example to enable molecular cloning, sequencing or simple detection of RNA. Conversely, qPCR may be used without RT-PCR, for example to quantify the copy number of a specific piece of DNA.
Nomenclature
The combined RT-PCR and qPCR technique has been described as quantitative RT-PCR[3] or real-time RT-PCR[4] (sometimes even called quantitative real-time RT-PCR[5]), has been variously abbreviated as qRT-PCR,[6] RT-qPCR,[7] RRT-PCR,[8] and rRT-PCR.[9] In order to avoid confusion, the following abbreviations will be used consistently throughout this article:
| Technique | Abbreviation |
|---|---|
| Polymerase chain reaction | PCR |
| Reverse transcription polymerase chain reaction | RT-PCR |
| Real-time polymerase chain reaction | qPCR |
| RT-PCR / qPCR combined technique | qRT-PCR |
Not all authors, especially earlier ones, use this convention and the reader should be cautious when following links. RT-PCR has been used to indicate both real-time PCR (qPCR) and reverse transcription PCR (RT-PCR).
History
Since its introduction in 1977, Northern blot has been used extensively for RNA quantification despite its shortcomings: (a) time-consuming technique, (b) requires a large quantity of RNA for detection, and (c) quantitatively inaccurate in the low abundance of RNA content.[10][11] However, since PCR was invented by Kary Mullis in 1983, RT PCR has since displaced Northern blot as the method of choice for RNA detection and quantification.[12]
RT-PCR has risen to become the benchmark technology for the detection and/or comparison of RNA levels for several reasons: (a) it does not require post PCR processing, (b) a wide range (>107-fold) of RNA abundance can be measured, and (c) it provides insight into both qualitative and quantitative data.[5] Due to its simplicity, specificity and sensitivity, RT-PCR is used in a wide range of applications from experiments as simple as quantification of yeast cells in wine to more complex uses as diagnostic tools for detecting infectious agents such as the avian flu virus and SARS-CoV-2.[13][14][15]
Principles
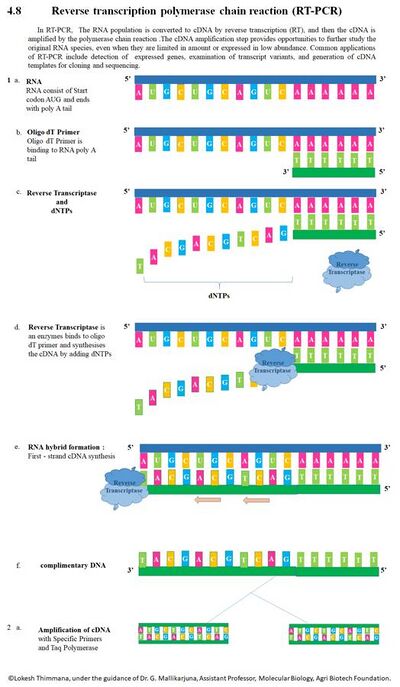
In RT-PCR, the RNA template is first converted into a complementary DNA (cDNA) using a reverse transcriptase (RT). The cDNA is then used as a template for exponential amplification using PCR. The use of RT-PCR for the detection of RNA transcript has revolutionized the study of gene expression in the following important ways:
- Made it theoretically possible to detect the transcripts of practically any gene[16]
- Enabled sample amplification and eliminated the need for abundant starting material required when using northern blot analysis[17][18]
- Provided tolerance for RNA degradation as long as the RNA spanning the primer is intact[17]
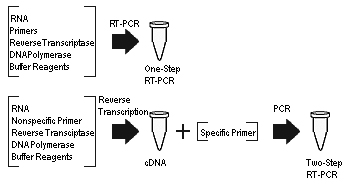
One-step RT-PCR vs two-step RT-PCR
The quantification of mRNA using RT-PCR can be achieved as either a one-step or a two-step reaction. The difference between the two approaches lies in the number of tubes used when performing the procedure. The two-step reaction requires that the reverse transcriptase reaction and PCR amplification be performed in separate tubes. The disadvantage of the two-step approach is susceptibility to contamination due to more frequent sample handling.[19] On the other hand, the entire reaction from cDNA synthesis to PCR amplification occurs in a single tube in the one-step approach. The one-step approach is thought to minimize experimental variation by containing all of the enzymatic reactions in a single environment. It eliminates the steps of pipetting cDNA product, which is labor-intensive and prone to contamination, to PCR reaction. The further use of inhibitor-tolerant polymerases, polymerase enhancers with an optimized one-step RT-PCR condition, supports the reverse transcription of the RNA from unpurified or crude samples, such as whole blood and serum.[20][21] However, the starting RNA templates are prone to degradation in the one-step approach, and the use of this approach is not recommended when repeated assays from the same sample is required. Additionally, the one-step approach is reported to be less accurate compared to the two-step approach. It is also the preferred method of analysis when using DNA binding dyes such as SYBR Green since the elimination of primer-dimers can be achieved through a simple change in the melting temperature. Nevertheless, the one-step approach is a relatively convenient solution for the rapid detection of target RNA directly in biosensing.[citation needed]
End-point RT-PCR vs real-time RT-PCR
Quantification of RT-PCR products can largely be divided into two categories: end-point and real-time.[22] The use of end-point RT-PCR is preferred for measuring gene expression changes in small number of samples, but the real-time RT-PCR has become the gold standard method for validating quantitative results obtained from array analyses or gene expression changes on a global scale.[23]
End-point RT-PCR
The measurement approaches of end-point RT-PCR requires the detection of gene expression levels by the use of fluorescent dyes like ethidium bromide,[24][25] P32 labeling of PCR products using phosphorimager,[26] or by scintillation counting.[18] End-point RT-PCR is commonly achieved using three different methods: relative, competitive and comparative.[27][28]
- Relative RT-PCR
- Relative quantifications of RT-PCR involves the co-amplification of an internal control simultaneously with the gene of interest. The internal control is used to normalize the samples. Once normalized, a direct comparison of relative transcript abundances across multiple samples of mRNA can be made. One precaution to note is that the internal control must be chosen so that it is not affected by the experimental treatment. The expression level should be constant across all samples and with the mRNA of interest for the results to be accurate and meaningful. Because the quantification of the results are analyzed by comparing the linear range of the target and control amplification, it is crucial to take into consideration the starting target molecules concentration and their amplification rate prior to starting the analysis. The results of the analysis are expressed as the ratios of gene signal to internal control signal, which the values can then be used for the comparison between the samples in the estimation of relative target RNA expression.[25][28][29]
- Competitive RT-PCR
- Competitive RT-PCR technique is used for absolute quantification. It involves the use of a synthetic “competitor” RNA that can be distinguished from the target RNA by a small difference in size or sequence. It is important for the design of the synthetic RNA be identical in sequence but slightly shorter than the target RNA for accurate results. Once designed and synthesized, a known amount of the competitor RNA is added to experimental samples and is co-amplified with the target using RT-PCR. Then, a concentration curve of the competitor RNA is produced and it is used to compare the RT-PCR signals produced from the endogenous transcripts to determine the amount of target present in the sample.[28][30]
- Comparative RT-PCR
- Comparative RT-PCR is similar to the competitive RT-PCR in that the target RNA competes for amplification reagents within a single reaction with an internal standard of unrelated sequence. Once the reaction is complete, the results are compared to an external standard curve to determine the target RNA concentration. In comparison to the relative and competitive quantification methods, comparative RT-PCR is considered to be the more convenient method to use since it does not require the investigator to perform a pilot experiment; in relative RT-PCR, the exponential amplification range of the mRNA must be predetermined and in competitive RT-PCR, a synthetic competitor RNA must be synthesized.[28][31][32][33][34]
Real-time RT-PCR
The emergence of novel fluorescent DNA labeling techniques in the past few years has enabled the analysis and detection of PCR products in real-time and has consequently led to the widespread adoption of real-time RT-PCR for the analysis of gene expression.[35] Not only is real-time RT-PCR now the method of choice for quantification of gene expression, it is also the preferred method of obtaining results from array analyses and gene expressions on a global scale.[36] Currently, there are four different fluorescent DNA probes available for the real-time RT-PCR detection of PCR products: SYBR Green, TaqMan, molecular beacons, and scorpion probes. All of these probes allow the detection of PCR products by generating a fluorescent signal. While the SYBR Green dye emits its fluorescent signal simply by binding to the double-stranded DNA in solution, the TaqMan probes', molecular beacons' and scorpions' generation of fluorescence depend on Förster Resonance Energy Transfer (FRET) coupling of the dye molecule and a quencher moiety to the oligonucleotide substrates.[37]
- SYBR Green
- When the SYBR Green binds to the double-stranded DNA of the PCR products, it will emit light upon excitation. The intensity of the fluorescence increases as the PCR products accumulate. This technique is easy to use since designing of probes is not necessary given lack of specificity of its binding. However, since the dye does not discriminate the double-stranded DNA from the PCR products and those from the primer-dimers, overestimation of the target concentration is a common problem. Where accurate quantification is an absolute necessity, further assay for the validation of results must be performed. Nevertheless, among the real-time RT-PCR product detection methods, SYBR Green is the most economical and easiest to use.[22][23]
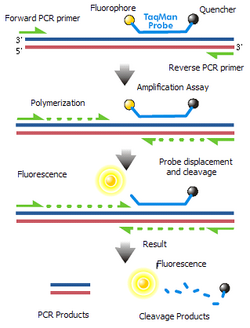
- TaqMan probes
- TaqMan probes are oligonucleotides that have a fluorescent probe attached to the 5' end and a quencher to the 3' end. During PCR amplification, these probes will hybridize to the target sequences located in the amplicon and as polymerase replicates the template with TaqMan bound, it also cleaves the fluorescent probe due to polymerase 5'- nuclease activity. Because the close proximity between the quench molecule and the fluorescent probe normally prevents fluorescence from being detected through FRET, the decoupling results in the increase of intensity of fluorescence proportional to the number of the probe cleavage cycles. Although well-designed TaqMan probes produce accurate real-time RT-PCR results, it is expensive and time-consuming to synthesize when separate probes must be made for each mRNA target analyzed.[22][16][38] Additionally, these probes are light sensitive and must be carefully frozen as aliquots to prevent degradation.
- Molecular beacon probes
- Similar to the TaqMan probes, molecular beacons also make use of FRET detection with fluorescent probes attached to the 5' end and a quencher attached to the 3' end of an oligonucleotide substrate. However, whereas the TaqMan fluorescent probes are cleaved during amplification, molecular beacon probes remain intact and rebind to a new target during each reaction cycle. When free in solution, the close proximity of the fluorescent probe and the quencher molecule prevents fluorescence through FRET. However, when molecular beacon probes hybridize to a target, the fluorescent dye and the quencher are separated resulting in the emittance of light upon excitation. As is with the TaqMan probes, molecular beacons are expensive to synthesize and require separate probes for each RNA target.[19]
- Scorpion probes
- The scorpion probes, like molecular beacons, will not be fluorescent active in an unhybridized state, again, due to the fluorescent probe on the 5' end being quenched by the moiety on the 3' end of an oligonucleotide. With Scorpions, however, the 3' end also contains sequence that is complementary to the extension product of the primer on the 5' end. When the Scorpion extension binds to its complement on the amplicon, the Scorpion structure opens, prevents FRET, and enables the fluorescent signal to be measured.[39]
- Multiplex probes
- TaqMan probes, molecular beacons, and scorpions allow the concurrent measurement of PCR products in a single tube. This is possible because each of the different fluorescent dyes can be associated with a specific emission spectra. Not only does the use of multiplex probes save time and effort without compromising test utility, its application in wide areas of research such as gene deletion analysis, mutation and polymorphism analysis, quantitative analysis, and RNA detection, make it an invaluable technique for laboratories of many discipline.[39][40][41]
Two strategies are commonly employed to quantify the results obtained by real-time RT-PCR; the standard curve method and the comparative threshold method.[42]
Application
The exponential amplification via reverse transcription polymerase chain reaction provides for a highly sensitive technique in which a very low copy number of RNA molecules can be detected. RT-PCR is widely used in the diagnosis of genetic diseases and, semiquantitatively, in the determination of the abundance of specific different RNA molecules within a cell or tissue as a measure of gene expression.
Research methods
RT-PCR is commonly used in research methods to measure gene expression. For example, Lin et al. used qRT-PCR to measure expression of Gal genes in yeast cells. First, Lin et al. engineered a mutation of a protein suspected to participate in the regulation of Gal genes. This mutation was hypothesized to selectively abolish Gal expression. To confirm this, gene expression levels of yeast cells containing this mutation were analyzed using qRT-PCR. The researchers were able to conclusively determine that the mutation of this regulatory protein reduced Gal expression.[43] Northern blot analysis is used to study the RNA's gene expression further.
Gene insertion
RT-PCR can also be very useful in the insertion of eukaryotic genes into prokaryotes. Because most eukaryotic genes contain introns, which are present in the genome but not in the mature mRNA, the cDNA generated from a RT-PCR reaction is the exact (without regard to the error-prone nature of reverse transcriptases) DNA sequence that would be directly translated into protein after transcription. When these genes are expressed in prokaryotic cells for the sake of protein production or purification, the RNA produced directly from transcription need not undergo splicing as the transcript contains only exons. (Prokaryotes, such as E. coli, lack the mRNA splicing mechanism of eukaryotes).
Genetic disease diagnosis
RT-PCR can be used to diagnose genetic disease such as Lesch–Nyhan syndrome. This genetic disease is caused by a malfunction in the HPRT1 gene, which clinically leads to the fatal uric acid urinary stone and symptoms similar to gout.[6][clarification needed] Analyzing a pregnant mother and a fetus for mRNA expression levels of HPRT1 will reveal if the mother is a carrier and if the fetus will likely to develop Lesch–Nyhan syndrome.[44]
Cancer detection
Scientists are working on ways to use RT-PCR in cancer detection to help improve prognosis, and monitor response to therapy. Circulating tumor cells produce unique mRNA transcripts depending on the type of cancer. The goal is to determine which mRNA transcripts serve as the best biomarkers for a particular cancer cell type and then analyze its expression levels with RT-PCR.[45]
RT-PCR is commonly used in studying the genomes of viruses whose genomes are composed of RNA, such as Influenzavirus A, retroviruses like HIV and SARS-CoV-2.[46]
Challenges
Despite its major advantages, RT-PCR is not without drawbacks. The exponential growth of the reverse transcribed complementary DNA (cDNA) during the multiple cycles of PCR produces inaccurate end point quantification due to the difficulty in maintaining linearity.[47] In order to provide accurate detection and quantification of RNA content in a sample, qRT-PCR was developed using fluorescence-based modification to monitor the amplification products during each cycle of PCR. The extreme sensitivity of the technique can be a double edged sword since even the slightest DNA contamination can lead to undesirable results.[48] A simple method for elimination of false positive results is to include anchors, or tags, to the 5' region of a gene specific primer.[49] Additionally, planning and design of quantification studies can be technically challenging due to the existence of numerous sources of variation including template concentration and amplification efficiency.[31] Spiking in a known quantity of RNA into a sample, adding a series of RNA dilutions generating a standard curve, and adding in a no template copy sample (no cDNA) may used as controls.
Protocol
RT-PCR can be carried out by the one-step RT-PCR protocol or the two-step RT-PCR protocol.
One-step RT-PCR
One-step RT-PCR subjects mRNA targets (up to 6 kb) to reverse transcription followed by PCR amplification in a single test tube. It is important to note that using intact, high quality RNA and a sequence-specific primer will produce the best results.
Once a one-step RT-PCR kit with a mix of reverse transcriptase, Taq DNA polymerase, and a proofreading polymerase is selected and all necessary materials and equipment are obtained a reaction mix is to be prepared. The reaction mix includes dNTPs, primers, template RNA, necessary enzymes, and a buffer solution. The reaction mix is added to a PCR tube for each reaction, followed by template RNA. The PCR tubes are then placed in a thermal cycler to begin cycling. In the first cycle, synthesis of cDNA occurs. The second cycle is the initial denaturation wherein reverse transcriptase is inactivated. The remaining 40-50 cycles are the amplification, which includes denaturation, annealing, and elongation. When amplification is complete, the RT-PCR products can be analyzed with gel electrophoresis.[50][51]
(PCR Applications Manual and Biotools)
Two-step RT-PCR
Two-step RT-PCR, as the name implies, occurs in two steps. First the reverse transcription and then the PCR. This method is more sensitive than the one-step method. Kits are also useful for two-step RT-PCR. Just as for one-step PCR, use only intact, high quality RNA for the best results. The primer for two-step PCR does not have to be sequence specific.
Step one
First combine template RNA, primer, dNTP mix, and nuclease-free water in a PCR tube. Then, add an RNase inhibitor and reverse transcriptase to the PCR tube. Next, place the PCR tube into a thermal cycler for one cycle wherein annealing, extending, and inactivating of reverse transcriptase occurs. Finally, proceed directly to step two which is PCR or store product on ice until PCR can be performed.
Step two
Add master mix which contains buffer, dNTP mix, MgCl2, Taq polymerase and nuclease-free water to each PCR tube. Then add the necessary primer to the tubes. Next, place the PCR tubes in a thermal cycler for 30 cycles of the amplification program. This includes: denaturation, annealing, and elongation. The products of RT-PCR can be analyzed with gel electrophoresis.[52]
Publication guidelines
Quantitative RT-PCR assay is considered to be the gold standard for measuring the number of copies of specific cDNA targets in a sample but it is poorly standardized.[53] As a result, while there are numerous publications utilizing the technique, many provide inadequate experimental detail and use unsuitable data analysis to draw inappropriate conclusions. Due to the inherent variability in the quality of any quantitative PCR data, not only do reviewers have a difficult time evaluating these manuscripts, but the studies also become impossible to replicate.[54] Recognizing the need for the standardization of the reporting of experimental conditions, the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE, pronounced mykee) guidelines have been published by an international consortium of academic scientists. The MIQE guidelines describe the minimum information necessary for evaluating quantitative PCR experiments that should be required for publication for encouraging better experimental practice and ensuring the relevance, accuracy, correct interpretation, and repeatability of quantitative PCR data.[55]
Besides reporting guidelines, the MIQE stresses the need to standardize the nomenclature associated with quantitative PCR to avoid confusion; for example, the abbreviation qPCR should be used for quantitative real-time PCR, while RT-qPCR should be used for reverse transcription-qPCR, and genes used for normalisation should be referred to as reference genes instead of housekeeping genes. It also proposes that commercially derived terms like TaqMan probes should not be used, but instead referred to as hydrolysis probes. Additionally, it is proposed that quantification cycle (Cq) be used to describe the PCR cycle used for quantification instead of threshold cycle (Ct), crossing point (Cp), and takeoff point (TOP), which refer to the same value but were coined by different manufacturers of real-time instruments.[53]
The guideline consists of the following elements: 1) experimental design, 2) sample, 3) nucleic acid extraction, 4) reverse transcription, 5) qPCR target information, 6) oligonucleotides, 7) protocol, 8) validation, and 9) data analysis. Specific items within each element carry a label of either E (essential) or D (desirable). Those labelled E are considered critical and indispensable while those labelled D are considered peripheral yet important for best-practices.[55]
References
- ↑ "Quantitative RT-PCR: pitfalls and potential". BioTechniques 26 (1): 112–22, 124–5. January 1999. doi:10.2144/99261rv01. PMID 9894600.
- ↑ Mackay, Ian (2007). Real-time PCR in Microbiology: From Diagnosis to Characterization. Norfolk, England: Caister Academic Press. pp. 440. ISBN 978-1-904455-18-9. https://archive.org/details/realtimepcrmicro00mack.
- ↑ Joyce C (2002). Quantitative RT-PCR. A review of current methodologies. Methods Mol. Biol.. 193. pp. 83–92. doi:10.1385/1-59259-283-X:083. ISBN 978-1-59259-283-8.
- ↑ "A duplex real-time RT-PCR assay for detecting H5N1 avian influenza virus and pandemic H1N1 influenza virus". Virol. J. 7: 113. 2010. doi:10.1186/1743-422X-7-113. PMID 20515509.
- ↑ 5.0 5.1 "Quantitative real-time RT-PCR--a perspective". J. Mol. Endocrinol. 34 (3): 597–601. June 2005. doi:10.1677/jme.1.01755. PMID 15956331.
- ↑ "qRT-PCR of Small RNAs". Plant Epigenetics. Methods in Molecular Biology. 631. 2010. pp. 109–22. doi:10.1007/978-1-60761-646-7_10. ISBN 978-1-60761-645-0.
- ↑ "A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines". Methods 50 (4): S1–5. April 2010. doi:10.1016/j.ymeth.2010.01.005. PMID 20215014.
- ↑ "Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes". J. Clin. Microbiol. 40 (9): 3256–60. September 2002. doi:10.1128/jcm.40.9.3256-3260.2002. PMID 12202562.
- ↑ "ACCELERATED EMERGENCY USE AUTHORIZATION (EUA) SUMMARY COVID-19 RT-PCR TEST (LABORATORY CORPORATION OF AMERICA)". https://www.fda.gov/media/136151/download.
- ↑ "Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes". Proc. Natl. Acad. Sci. U.S.A. 74 (12): 5350–4. December 1977. doi:10.1073/pnas.74.12.5350. PMID 414220. Bibcode: 1977PNAS...74.5350A.
- ↑ "Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues". Nat Protoc 4 (1): 37–43. 2009. doi:10.1038/nprot.2008.216. PMID 19131955.
- ↑ Bustin SA (October 2000). "Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays". J. Mol. Endocrinol. 25 (2): 169–93. doi:10.1677/jme.0.0250169. PMID 11013345.
- ↑ "Real-time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeasts in wine". Appl. Environ. Microbiol. 72 (11): 7148–55. November 2006. doi:10.1128/AEM.00388-06. PMID 17088381. Bibcode: 2006ApEnM..72.7148H.
- ↑ "Validated RealTime reverse transcriptase PCR methods for the diagnosis and pathotyping of Eurasian H7 avian influenza viruses". Influenza and Other Respiratory Viruses 3 (4): 151–64. July 2009. doi:10.1111/j.1750-2659.2009.00083.x. PMID 19627372.
- ↑ Mission summary: WHO Field Visit to Wuhan, China 20–21 January 2020: https://www.who.int/china/news/detail/22-01-2020-field-visit-wuhan-china-jan-2020
- ↑ 16.0 16.1 "Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes". Curr. Genomics 8 (4): 234–51. June 2007. doi:10.2174/138920207781386960. PMID 18645596.
- ↑ 17.0 17.1 Bustin SA (August 2002). "Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems". J. Mol. Endocrinol. 29 (1): 23–39. doi:10.1677/jme.0.0290023. PMID 12200227.
- ↑ 18.0 18.1 "Quantitative RT-PCR: limits and accuracy". BioTechniques 21 (2): 280–5. August 1996. doi:10.2144/96212rr01. PMID 8862813.
- ↑ 19.0 19.1 "Real-time PCR for mRNA quantitation". BioTechniques 39 (1): 75–85. July 2005. doi:10.2144/05391rv01. PMID 16060372.
- ↑ Li, Lang; He, Jian-an; Wang, Wei; Xia, Yun; Song, Li; Chen, Ze-han; Zuo, Hang-zhi; Tan, Xuan-Ping et al. (2019-08-01). "Development of a direct reverse-transcription quantitative PCR (dirRT-qPCR) assay for clinical Zika diagnosis" (in en). International Journal of Infectious Diseases 85: 167–174. doi:10.1016/j.ijid.2019.06.007. ISSN 1201-9712. PMID 31202908. https://www.ijidonline.com/article/S1201-9712(19)30252-8/abstract.
- ↑ Bachofen, Claudia; Willoughby, Kim; Zadoks, Ruth; Burr, Paul; Mellor, Dominic; Russell, George C. (2013-06-01). "Direct RT-PCR from serum enables fast and cost-effective phylogenetic analysis of bovine viral diarrhoea virus". Journal of Virological Methods 190 (1): 1–3. doi:10.1016/j.jviromet.2013.03.015. ISSN 0166-0934. PMID 23541784.
- ↑ 22.0 22.1 22.2 "Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods". Anal. Biochem. 285 (2): 194–204. October 2000. doi:10.1006/abio.2000.4753. PMID 11017702.
- ↑ 23.0 23.1 "Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR". J Mol Diagn 3 (1): 26–31. February 2001. doi:10.1016/S1525-1578(10)60646-0. PMID 11227069.
- ↑ "Detection of African horse sickness virus by reverse transcription-PCR". J. Clin. Microbiol. 32 (3): 697–700. March 1994. doi:10.1128/JCM.32.3.697-700.1994. PMID 8195381.
- ↑ 25.0 25.1 Minton AP (April 1995). "Confinement as a determinant of macromolecular structure and reactivity. II. Effects of weakly attractive interactions between confined macrosolutes and confining structures". Biophys. J. 68 (4): 1311–22. doi:10.1016/S0006-3495(95)80304-8. PMID 7787020. Bibcode: 1995BpJ....68.1311M.
- ↑ "Functional analysis of the single Est1/Ebs1 homologue in Kluyveromyces lactis reveals roles in both telomere maintenance and rapamycin resistance". Eukaryotic Cell 11 (7): 932–42. July 2012. doi:10.1128/EC.05319-11. PMID 22544908.
- ↑ "Analyzing real-time PCR data by the comparative C(T) method". Nat Protoc 3 (6): 1101–8. 2008. doi:10.1038/nprot.2008.73. PMID 18546601.
- ↑ 28.0 28.1 28.2 28.3 Tang, Yi-Wei (2012-09-13), Advanced Techniques in Diagnostic Microbiology, Springer, ISBN 978-1461439691
- ↑ "The use of the PCR to quantitate gene expression". Genome Research 3 (6): S123–35. June 1994. doi:10.1101/gr.3.6.s123. PMID 7522722.
- ↑ "Quantification of mRNA using competitive RT-PCR with standard-curve methodology". BioTechniques 21 (5): 862–6. November 1996. doi:10.2144/96215st04. PMID 8922627.
- ↑ 31.0 31.1 "Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data". Neurosci. Lett. 339 (1): 62–6. March 2003. doi:10.1016/S0304-3940(02)01423-4. PMID 12618301.
- ↑ "The inherent quantitative capacity of the reverse transcription-polymerase chain reaction". Anal. Biochem. 266 (2): 181–91. January 1999. doi:10.1006/abio.1998.2913. PMID 9888974.
- ↑ King N (2010). "The use of comparative quantitative RT-PCR to investigate the effect of cysteine incubation on GPx1 expression in freshly isolated cardiomyocytes". RT-PCR Protocols. Methods in Molecular Biology. 630. pp. 215–32. doi:10.1007/978-1-60761-629-0_14. ISBN 978-1-60761-628-3.
- ↑ "A reverse transcription comparative real-time PCR method for quantitative detection of angiogenic growth factors in head and neck cancer patients". Clin. Biochem. 35 (8): 591–6. November 2002. doi:10.1016/S0009-9120(02)00403-4. PMID 12498992.
- ↑ Lu, Rou-Jian; Zhao, Li; Huang, Bao-Ying; Ye, Fei; Wang, Wen-Ling; Tan, Wen-Jie (5 September 2021). "Real-time reverse transcription-polymerase chain reaction assay panel for the detection of severe acute respiratory syndrome coronavirus 2 and its variants". Chinese Medical Journal 134 (17): 2048–2053. doi:10.1097/CM9.0000000000001687. PMID 34402479. PMC 8439998. https://journals.lww.com/cmj/Fulltext/2021/09050/Real_time_reverse_transcription_polymerase_chain.5.aspx. Retrieved 17 February 2023.
- ↑ Jawerth, Nicole (27 March 2020). "How is the COVID-19 Virus Detected using real time reverse transcription–polymerase chain reaction?" (in en). https://www.iaea.org/newscenter/news/how-is-the-covid-19-virus-detected-using-real-time-rt-pcr.
- ↑ Holden, M. J.; Wang, L. (2008). "Quantitative Real-Time PCR: Fluorescent Probe Options and Issues". Standardization and Quality Assurance in Fluorescence Measurements II. Springer Series on Fluorescence. 6. pp. 489. doi:10.1007/4243_2008_046. ISBN 978-3-540-70570-3. https://zenodo.org/record/1232956.
- ↑ "TaqMan reverse transcription polymerase chain reaction for the detection of Japanese encephalitis virus". J. Vet. Sci. 5 (4): 345–51. December 2004. doi:10.4142/jvs.2004.5.4.345. PMID 15613819.
- ↑ 39.0 39.1 "Detection and quantification of gene expression in environmental bacteriology". Appl. Environ. Microbiol. 70 (7): 3795–806. July 2004. doi:10.1128/AEM.70.7.3795-3806.2004. PMID 15240248. Bibcode: 2004ApEnM..70.3795S.
- ↑ "Molecular diagnosis of medical viruses". Curr Issues Mol Biol 9 (2): 87–102. July 2007. PMID 17489437.
- ↑ "Multiplex PCR: optimization and application in diagnostic virology". Clin. Microbiol. Rev. 13 (4): 559–70. October 2000. doi:10.1128/cmr.13.4.559-570.2000. PMID 11023957.
- ↑ Bustin SA (July 2005). "Real-time, fluorescence-based quantitative PCR: a snapshot of current procedures and preferences". Expert Rev. Mol. Diagn. 5 (4): 493–8. doi:10.1586/14737159.5.4.493. PMID 16013967.
- ↑ "Analysis of Gal4-directed transcription activation using Tra1 mutants selectively defective for interaction with Gal4". Proc. Natl. Acad. Sci. U.S.A. 109 (6): 1997–2002. February 2012. doi:10.1073/pnas.1116340109. PMID 22308403. Bibcode: 2012PNAS..109.1997L.
- ↑ "Carrier and prenatal diagnosis of Lesch-Nyhan disease due to a defect in HPRT gene expression regulation". Gene 511 (2): 306–7. December 2012. doi:10.1016/j.gene.2012.09.121. PMID 23046577.
- ↑ "Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers". Clin. Chem. 53 (7): 1206–15. July 2007. doi:10.1373/clinchem.2006.081828. PMID 17525108.
- ↑ "Coronavirus: il viaggio dei test". https://www.iss.it/web/guest/primo-piano/-/asset_publisher/o4oGR9qmvUz9/content/id/5269706.
- ↑ Shiao YH (December 2003). "A new reverse transcription-polymerase chain reaction method for accurate quantification". BMC Biotechnol. 3: 22. doi:10.1186/1472-6750-3-22. PMID 14664723.
- ↑ "Reverse transcription-PCR analysis of the regulation of the manganese peroxidase gene family". Appl. Environ. Microbiol. 64 (2): 569–74. February 1998. doi:10.1128/AEM.64.2.569-574.1998. PMID 9464395. Bibcode: 1998ApEnM..64..569G.
- ↑ Martel, Fatima; Dirk Grundemann; Edgar Schöig (2002-03-31). "A simple method for elimination of false positive results in RT-PCR". J Biochem Mol Biol 35 (2): 248–250. doi:10.5483/BMBRep.2002.35.2.248. PMID 12297038.
- ↑ "High Transcript Tools OneStep Kit". Biotools. http://www.biotools.eu/reverse_transcription.html.
- ↑ Degen, Hans-Joachim; Deufel, Annette; Eisel, Doris; Grünewald-Janho, Stefanie; Keesey, Joe (2006). PCR Applications Manual (3 ed.). Roche Diagnostics. pp. 135–137. http://www.roche-applied-science.com/publications/print_mat/pcr_application_manual_3rd_edition.pdf.
- ↑ "RT-PCR Two-Step Protocol". MIT. http://ocw.mit.edu/courses/biology/7-16-experimental-molecular-biology-biotechnology-ii-spring-2005/labs/rt_pcr_2step.pdf.
- ↑ 53.0 53.1 "www.microarrays.ca". http://www.microarrays.ca/about/reviews/Bustin_Review_July2009.pdf.
- ↑ Bustin SA (April 2010). "Why the need for qPCR publication guidelines?--The case for MIQE". Methods 50 (4): 217–26. doi:10.1016/j.ymeth.2009.12.006. PMID 20025972.
- ↑ 55.0 55.1 "The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments". Clin. Chem. 55 (4): 611–22. April 2009. doi:10.1373/clinchem.2008.112797. PMID 19246619.
External links
- RT-PCR protocols from Penn state University
- Database of validated PCR primer sets (website critique)
- Animation to illustrate RT-PCR procedure, from Cold Spring Harbor Laboratory
- The Reference in qPCR – an Academic & Industrial Information Platform
- Top 5 Government Rt Pcr Centres in Mumbai
fr:RT-PCR#Après transcription inverse
 |