Biology:X-inactivation


1.Early stage embryonic cell of a female human
2.Maternal X chromosome
3.Paternal X chromosome
4.Mitosis and random X-chromosome inactivation event
5.Paternal chromosome is randomly inactivated in one daughter cell, maternal chromosome is inactivated in the other
6.Paternal chromosome is randomly inactivated in both daughter cells
7.Maternal chromosome is randomly inactivated in both daughter cells
8.Three possible random combination outcomes
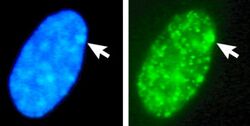
Left: DNA (DAPI)-stained nucleus. Arrow indicates the location of Barr body(Xi). Right: DNA associated histones protein detected

X-inactivation (also called Lyonization, after English geneticist Mary Lyon) is a process by which one of the copies of the X chromosome is inactivated in therian female mammals. The inactive X chromosome is silenced by being packaged into a transcriptionally inactive structure called heterochromatin. As nearly all female mammals have two X chromosomes, X-inactivation prevents them from having twice as many X chromosome gene products as males, who only possess a single copy of the X chromosome (see dosage compensation).
The choice of which X chromosome will be inactivated in a particular embryonic cell is random in placental mammals such as humans, but once an X chromosome is inactivated it will remain inactive throughout the lifetime of the cell and its descendants in the organism (its cell line). The result is that the choice of inactivated X chromosome in all the cells of the organism is a random distribution, often with about half the cells having the paternal X chromosome inactivated and half with an inactivated maternal X chromosome; but commonly, X-inactivation is unevenly distributed across the cell lines within one organism (skewed X-inactivation).
Unlike the random X-inactivation in placental mammals, inactivation in marsupials applies exclusively to the paternally-derived X chromosome.
Mechanism
Cycle of X-chromosome activation in rodents
The paragraphs below have to do only with rodents and do not reflect XI in the majority of mammals. X-inactivation is part of the activation cycle of the X chromosome throughout the female life. The egg and the fertilized zygote initially use maternal transcripts, and the whole embryonic genome is silenced until zygotic genome activation. Thereafter, all mouse cells undergo an early, imprinted inactivation of the paternally-derived X chromosome in 4–8 cell stage embryos.[3][4][5][6] The extraembryonic tissues (which give rise to the placenta and other tissues supporting the embryo) retain this early imprinted inactivation, and thus only the maternal X chromosome is active in these tissues.
In the early blastocyst, this initial, imprinted X-inactivation is reversed in the cells of the inner cell mass (which give rise to the embryo), and in these cells both X chromosomes become active again. Each of these cells then independently and randomly inactivates one copy of the X chromosome.[5] This inactivation event is irreversible during the lifetime of the individual, with the exception of the germline. In the female germline before meiotic entry, X-inactivation is reversed, so that after meiosis all haploid oocytes contain a single active X chromosome.
Overview
The Xi marks the inactive, Xa the active X chromosome. XP denotes the paternal, and XM to denotes the maternal X chromosome. When the egg (carrying XM), is fertilized by a sperm (carrying a Y or an XP) a diploid zygote forms. From zygote, through adult stage, to the next generation of eggs, the X chromosome undergoes the following changes:
- XiP XiM zygote → undergoing zygotic genome activation, leading to:
- XaP XaM → undergoing imprinted (paternal) X-inactivation, leading to:
- XiP XaM → undergoing X-activation in the early blastocyst stage, leading to:
- XaP XaM → undergoing random X-inactivation in the embryonic lineage (inner cell mass) in the blastocyst stage, leading to:
- XiP XaM OR XaP XiM → undergoing X-reactivation in primordial germ cells before meiosis, leading to:
- XaM XaP diploid germ cells in meiotic arrest. As the meiosis I only completes with ovulation, human germ cells exist in this stage from the first weeks of development until puberty. The completion of meiosis leads to:
- XaM AND XaP haploid germ cells (eggs).
The X activation cycle has been best studied in mice, but there are multiple studies in humans. As most of the evidence is coming from mice, the above scheme represents the events in mice. The completion of the meiosis is simplified here for clarity. Steps 1–4 can be studied in in vitro fertilized embryos, and in differentiating stem cells; X-reactivation happens in the developing embryo, and subsequent (6–7) steps inside the female body, therefore much harder to study.
Timing
The timing of each process depends on the species, and in many cases the precise time is actively debated. [The whole part of the human timing of X-inactivation in this table is highly questionable and should be removed until properly substantiated by empirical data]
| Process | Mouse | Human | |
| 1 | Zygotic genome activation | 2–4 cell stage[7] | 2–8 cell stage[7] |
| 2 | Imprinted (paternal) X-inactivation | 4–8 cell stage[6][8] | Unclear if it takes place in humans[9] |
| 3 | X-activation | Early blastocyst stage | Early blastocyst stage |
| 4 | Random X-inactivation in the embryonic lineage (inner cell mass) | Late blastocyst stage | Late blastocyst stage, after implantation[9] |
| 5 | X-reactivation in primordial germ cells before meiosis | From before developmental week 4 up to week 14[10][11] |
Inheritance of inactivation status across cell generations
The descendants of each cell which inactivated a particular X chromosome will also inactivate that same chromosome. This phenomenon, which can be observed in the coloration of tortoiseshell cats when females are heterozygous for the X-linked pigment gene, should not be confused with mosaicism, which is a term that specifically refers to differences in the genotype of various cell populations in the same individual; X-inactivation, which is an epigenetic change that results in a different phenotype, is not a change at the genotypic level. For an individual cell or lineage the inactivation is therefore skewed or 'non-random', and this can give rise to mild symptoms in female 'carriers' of X-linked genetic disorders.[12]
Selection of one active X chromosome
Typical females possess two X chromosomes, and in any given cell one chromosome will be active (designated as Xa) and one will be inactive (Xi). However, studies of individuals with extra copies of the X chromosome show that in cells with more than two X chromosomes there is still only one Xa, and all the remaining X chromosomes are inactivated. This indicates that the default state of the X chromosome in females is inactivation, but one X chromosome is always selected to remain active.
It is understood that X-chromosome inactivation is a random process, occurring at about the time of gastrulation in the epiblast (cells that will give rise to the embryo). The maternal and paternal X chromosomes have an equal probability of inactivation. This would suggest that women would be expected to suffer from X-linked disorders approximately 50% as often as men (because women have two X chromosomes, while men have only one); however, in actuality, the occurrence of these disorders in females is much lower than that. One explanation for this disparity is that 12–20% [13] of genes on the inactivated X chromosome remain expressed, thus providing women with added protection against defective genes coded by the X-chromosome. Some[who?] suggest that this disparity must be evidence of preferential (non-random) inactivation. Preferential inactivation of the paternal X-chromosome occurs in both marsupials and in cell lineages that form the membranes surrounding the embryo,[14] whereas in placental mammals either the maternally or the paternally derived X-chromosome may be inactivated in different cell lines.[15]
The time period for X-chromosome inactivation explains this disparity. Inactivation occurs in the epiblast during gastrulation, which gives rise to the embryo.[16] Inactivation occurs on a cellular level, resulting in a mosaic expression, in which patches of cells have an inactive maternal X-chromosome, while other patches have an inactive paternal X-chromosome. For example, a female heterozygous for haemophilia (an X-linked disease) would have about half of her liver cells functioning properly, which is typically enough to ensure normal blood clotting.[17][18] Chance could result in significantly more dysfunctional cells; however, such statistical extremes are unlikely. Genetic differences on the chromosome may also render one X-chromosome more likely to undergo inactivation. Also, if one X-chromosome has a mutation hindering its growth or rendering it non viable, cells which randomly inactivated that X will have a selective advantage over cells which randomly inactivated the normal allele. Thus, although inactivation is initially random, cells that inactivate a normal allele (leaving the mutated allele active) will eventually be overgrown and replaced by functionally normal cells in which nearly all have the same X-chromosome activated.[17]
It is hypothesized that there is an autosomally-encoded 'blocking factor' which binds to the X chromosome and prevents its inactivation.[19] The model postulates that there is a limiting blocking factor, so once the available blocking factor molecule binds to one X chromosome the remaining X chromosome(s) are not protected from inactivation. This model is supported by the existence of a single Xa in cells with many X chromosomes and by the existence of two active X chromosomes in cell lines with twice the normal number of autosomes.[20]
Sequences at the X inactivation center (XIC), present on the X chromosome, control the silencing of the X chromosome. The hypothetical blocking factor is predicted to bind to sequences within the XIC.
Expression of X-linked disorders in heterozygous females
The effect of female X heterozygosity is apparent in some localized traits, such as the unique coat pattern of a calico cat. It can be more difficult, however, to fully understand the expression of un-localized traits in these females, such as the expression of disease.
Since males only have one copy of the X chromosome, all expressed X-chromosomal genes (or alleles, in the case of multiple variant forms for a given gene in the population) are located on that copy of the chromosome. Females, however, will primarily express the genes or alleles located on the X-chromosomal copy that remains active. Considering the situation for one gene or multiple genes causing individual differences in a particular phenotype (i.e., causing variation observed in the population for that phenotype), in homozygous females it doesn't particularly matter which copy of the chromosome is inactivated, as the alleles on both copies are the same. However, in females that are heterozygous at the causal genes, the inactivation of one copy of the chromosome over the other can have a direct impact on their phenotypic value. Because of this phenomenon, there is an observed increase in phenotypic variation in females that are heterozygous at the involved gene or genes than in females that are homozygous at that gene or those genes.[21] There are many different ways in which the phenotypic variation can play out. In many cases, heterozygous females may be asymptomatic or only present minor symptoms of a given disorder, such as with X-linked adrenoleukodystrophy.[22]
The differentiation of phenotype in heterozygous females is furthered by the presence of X-inactivation skewing. Typically, each X-chromosome is silenced in half of the cells, but this process is skewed when preferential inactivation of a chromosome occurs. It is thought that skewing happens either by chance or by a physical characteristic of a chromosome that may cause it to be silenced more or less often, such as an unfavorable mutation.[23][24]
On average, each X chromosome is inactivated in half of the cells, although 5-20% of women display X-inactivation skewing.[23] In cases where skewing is present, a broad range of symptom expression can occur, resulting in expression varying from minor to severe depending on the skewing proportion. An extreme case of this was seen where monozygotic female twins had extreme variance in expression of Menkes disease (an X-linked disorder) resulting in the death of one twin while the other remained asymptomatic.[25]
It is thought that X-inactivation skewing could be caused by issues in the mechanism that causes inactivation, or by issues in the chromosome itself.[23][24] However, the link between phenotype and skewing is still being questioned, and should be examined on a case-by-case basis. A study looking at both symptomatic and asymptomatic females who were heterozygous for Duchenne and Becker muscular dystrophies (DMD) found no apparent link between transcript expression and skewed X-Inactivation. The study suggests that both mechanisms are independently regulated, and there are other unknown factors at play.[26]
Chromosomal component
The X-inactivation center (or simply XIC) on the X chromosome is necessary and sufficient to cause X-inactivation. Chromosomal translocations which place the XIC on an autosome lead to inactivation of the autosome, and X chromosomes lacking the XIC are not inactivated.[27][28]
The XIC contains four non-translated RNA genes, Xist, Tsix, Jpx and Ftx, which are involved in X-inactivation. The XIC also contains binding sites for both known and unknown regulatory proteins.[29]
Xist and Tsix RNAs
The X-inactive specific transcript (Xist) gene encodes a large non-coding RNA that is responsible for mediating the specific silencing of the X chromosome from which it is transcribed.[30] The inactive X chromosome is coated by Xist RNA,[31] whereas the Xa is not (See Figure to the right). X chromosomes that lack the Xist gene cannot be inactivated.[32] Artificially placing and expressing the Xist gene on another chromosome leads to silencing of that chromosome.[33][27]
Prior to inactivation, both X chromosomes weakly express Xist RNA from the Xist gene. During the inactivation process, the future Xa ceases to express Xist, whereas the future Xi dramatically increases Xist RNA production. On the future Xi, the Xist RNA progressively coats the chromosome, spreading out from the XIC;[33] the Xist RNA does not localize to the Xa. The silencing of genes along the Xi occurs soon after coating by Xist RNA.
Like Xist, the Tsix gene encodes a large RNA which is not believed to encode a protein. The Tsix RNA is transcribed antisense to Xist, meaning that the Tsix gene overlaps the Xist gene and is transcribed on the opposite strand of DNA from the Xist gene.[28] Tsix is a negative regulator of Xist; X chromosomes lacking Tsix expression (and thus having high levels of Xist transcription) are inactivated much more frequently than normal chromosomes.
Like Xist, prior to inactivation, both X chromosomes weakly express Tsix RNA from the Tsix gene. Upon the onset of X-inactivation, the future Xi ceases to express Tsix RNA (and increases Xist expression), whereas Xa continues to express Tsix for several days.
Rep A is a long non coding RNA that works with another long non coding RNA, Xist, for X inactivation. Rep A inhibits the function of Tsix, the antisense of Xist, in conjunction with eliminating expression of Xite. It promotes methylation of the Tsix region by attracting PRC2 and thus inactivating one of the X chromosomes.[29]
Silencing
The inactive X chromosome does not express the majority of its genes, unlike the active X chromosome. This is due to the silencing of the Xi by repressive heterochromatin, which compacts the Xi DNA and prevents the expression of most genes.
Compared to the Xa, the Xi has high levels of DNA methylation, low levels of histone acetylation, low levels of histone H3 lysine-4 methylation, and high levels of histone H3 lysine-9 methylation and H3 lysine-27 methylation mark which is placed by the PRC2 complex recruited by Xist, all of which are associated with gene silencing.[34] PRC2 regulates chromatin compaction and chromatin remodeling in several processes including the DNA damage response.[35] Additionally, a histone variant called macroH2A (H2AFY) is exclusively found on nucleosomes along the Xi.[36][37]
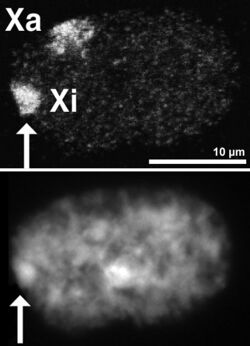
Barr bodies
DNA packaged in heterochromatin, such as the Xi, is more condensed than DNA packaged in euchromatin, such as the Xa. The inactive X forms a discrete body within the nucleus called a Barr body.[38] The Barr body is generally located on the periphery of the nucleus, is late replicating within the cell cycle, and, as it contains the Xi, contains heterochromatin modifications and the Xist RNA.
Expressed genes on the inactive X chromosome
A fraction of the genes along the X chromosome escape inactivation on the Xi. The Xist gene is expressed at high levels on the Xi and is not expressed on the Xa.[39] Many other genes escape inactivation; some are expressed equally from the Xa and Xi, and others, while expressed from both chromosomes, are still predominantly expressed from the Xa.[40][41][42] Up to one quarter of genes on the human Xi are capable of escape.[40] Studies in the mouse suggest that in any given cell type, 3% to 15% of genes escape inactivation, and that escaping gene identity varies between tissues.[41][42]
Many of the genes which escape inactivation are present along regions of the X chromosome which, unlike the majority of the X chromosome, contain genes also present on the Y chromosome. These regions are termed pseudoautosomal regions, as individuals of either sex will receive two copies of every gene in these regions (like an autosome), unlike the majority of genes along the sex chromosomes. Since individuals of either sex will receive two copies of every gene in a pseudoautosomal region, no dosage compensation is needed for females, so it is postulated that these regions of DNA have evolved mechanisms to escape X-inactivation. The genes of pseudoautosomal regions of the Xi do not have the typical modifications of the Xi and have little Xist RNA bound.
The existence of genes along the inactive X which are not silenced explains the defects in humans with abnormal numbers of the X chromosome, such as Turner syndrome (X0, caused by SHOX gene[43]) or Klinefelter syndrome (XXY). Theoretically, X-inactivation should eliminate the differences in gene dosage between affected individuals and individuals with a normal chromosome complement. In affected individuals, however, X-inactivation is incomplete and the dosage of these non-silenced genes will differ as they escape X-inactivation, similar to an autosomal aneuploidy.
The precise mechanisms that control escape from X-inactivation are not known, but silenced and escape regions have been shown to have distinct chromatin marks.[41][44] It has been suggested that escape from X-inactivation might be mediated by expression of long non-coding RNA (lncRNA) within the escaping chromosomal domains.[2]
Uses in experimental biology
Stanley Michael Gartler used X-chromosome inactivation to demonstrate the clonal origin of cancers. Examining normal tissues and tumors from females heterozygous for isoenzymes of the sex-linked G6PD gene demonstrated that tumor cells from such individuals express only one form of G6PD, whereas normal tissues are composed of a nearly equal mixture of cells expressing the two different phenotypes. This pattern suggests that a single cell, and not a population, grows into a cancer.[45] However, this pattern has been proven wrong for many cancer types, suggesting that some cancers may be polyclonal in origin.[46]
Besides, measuring the methylation (inactivation) status of the polymorphic human androgen receptor (HUMARA) located on X-chromosome is considered the most accurate method to assess clonality in female cancer biopsies.[47] A great variety of tumors was tested by this method, some, such as renal cell carcinoma,[48] found monoclonal while others (e.g. mesothelioma[49]) were reported polyclonal.
Researchers have also investigated using X-chromosome inactivation to silence the activity of autosomal chromosomes. For example, Jiang et al. inserted a copy of the Xist gene into one copy of chromosome 21 in stem cells derived from an individual with trisomy 21 (Down syndrome).[50] The inserted Xist gene induces Barr body formation, triggers stable heterochromatin modifications, and silences most of the genes on the extra copy of chromosome 21. In these modified stem cells, the Xist-mediated gene silencing seems to reverse some of the defects associated with Down syndrome.
History
In 1959 Susumu Ohno showed that the two X chromosomes of mammals were different: one appeared similar to the autosomes; the other was condensed and heterochromatic.[51] This finding suggested, independently to two groups of investigators, that one of the X chromosomes underwent inactivation.
In 1961, Mary Lyon proposed the random inactivation of one female X chromosome to explain the mottled phenotype of female mice heterozygous for coat color genes.[52] The Lyon hypothesis also accounted for the findings that one copy of the X chromosome in female cells was highly condensed, and that mice with only one copy of the X chromosome developed as infertile females. This suggested[53] to Ernest Beutler, studying heterozygous females for glucose-6-phosphate dehydrogenase (G6PD) deficiency, that there were two red cell populations of erythrocytes in such heterozygotes: deficient cells and normal cells,[54] depending on whether the inactivated X chromosome (in the nucleus of the red cell's precursor cell) contains the normal or defective G6PD allele.
See also
- Sex-determination system
- Dosage compensation
- Barr body
- Heterochromatin
- Epigenetics
- Skewed X-inactivation
- Developmental disorders thought to be related to X-inactivation:
References
- ↑ "Normal histone modifications on the inactive X chromosome in ICF and Rett syndrome cells: implications for methyl-CpG binding proteins". BMC Biology 2: 21. September 2004. doi:10.1186/1741-7007-2-21. PMID 15377381.
- ↑ 2.0 2.1 "Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse". BMC Genomics 11: 614. November 2010. doi:10.1186/1471-2164-11-614. PMID 21047393.
- ↑ "Preferential inactivation of the paternally derived X chromosome in the extra embryonic membranes of the mouse". Nature 256 (5519): 640–2. August 1975. doi:10.1038/256640a0. PMID 1152998. Bibcode: 1975Natur.256..640T.
- ↑ "Silence of the fathers: early X inactivation". BioEssays 26 (8): 821–4. August 2004. doi:10.1002/bies.20082. PMID 15273983. http://www3.interscience.wiley.com/cgi-bin/fulltext/109565168/PDFSTART.[|permanent dead link|dead link}}]
- ↑ 5.0 5.1 "Epigenetic dynamics of imprinted X inactivation during early mouse development". Science 303 (5658): 644–9. January 2004. doi:10.1126/science.1092727. PMID 14671313. Bibcode: 2004Sci...303..644O.
- ↑ 6.0 6.1 "Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells". Science 343 (6167): 193–6. January 2014. doi:10.1126/science.1245316. PMID 24408435. Bibcode: 2014Sci...343..193D.
- ↑ 7.0 7.1 "Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing" (in En). Nature 500 (7464): 593–7. August 2013. doi:10.1038/nature12364. PMID 23892778. Bibcode: 2013Natur.500..593X.
- ↑ "Xist-dependent imprinted X inactivation and the early developmental consequences of its failure" (in En). Nature Structural & Molecular Biology 24 (3): 226–233. March 2017. doi:10.1038/nsmb.3365. PMID 28134930.
- ↑ 9.0 9.1 "X chromosome regulation: diverse patterns in development, tissues and disease" (in En). Nature Reviews. Genetics 15 (6): 367–78. June 2014. doi:10.1038/nrg3687. PMID 24733023.
- ↑ "Parental haplotype-specific single-cell transcriptomics reveal incomplete epigenetic reprogramming in human female germ cells" (in En). Nature Communications 9 (1): 1873. May 2018. doi:10.1038/s41467-018-04215-7. PMID 29760424. Bibcode: 2018NatCo...9.1873V.
- ↑ "The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells". Cell 161 (6): 1437–52. June 2015. doi:10.1016/j.cell.2015.05.015. PMID 26046443.
- ↑ "X inactivation in females with X-linked disease". The New England Journal of Medicine 338 (5): 325–8. January 1998. doi:10.1056/NEJM199801293380611. PMID 9445416.
- ↑ "Derivation of consensus inactivation status for X-linked genes from genome-wide studies". Biology of Sex Differences 6 (35): 35. 30 December 2015. doi:10.1186/s13293-015-0053-7. PMID 26719789.
- ↑ "Mammals that break the rules: genetics of marsupials and monotremes". Annual Review of Genetics 30: 233–60. 1996. doi:10.1146/annurev.genet.30.1.233. PMID 8982455.
- ↑ "X-chromosome inactivation and developmental patterns in mammals". Biological Reviews of the Cambridge Philosophical Society 47 (1): 1–35. January 1972. doi:10.1111/j.1469-185X.1972.tb00969.x. PMID 4554151.
- ↑ "X chromosome inactivation in human cells". The Biomedical & Life Sciences Collection (Henry Stewart Talks, Ltd): 1–54. 2010. http://hstalks.com/main/view_talk.php?t=1676&r=494&c=252. Retrieved 15 December 2013.
- ↑ 17.0 17.1 Gartler, Stanley M; Goldman, Michael A (2001). "X-Chromosome Inactivation". Encyclopedia of Life Sciences (Nature Publishing Group): 1–2. http://web.udl.es/usuaris/e4650869/docencia/segoncicle/genclin98/recursos_classe_(pdf)/revisionsPDF/XChromoInac.pdf.
- ↑ "Sex-differential selection and the evolution of X inactivation strategies". PLOS Genetics 9 (4): e1003440. April 2013. doi:10.1371/journal.pgen.1003440. PMID 23637618.
- ↑ Avner, Philip; Heard, Edith (January 2001). "X-chromosome inactivation: counting, choice and initiation" (in en). Nature Reviews Genetics 2 (1): 59–67. doi:10.1038/35047580. ISSN 1471-0064. PMID 11253071. https://www.nature.com/articles/35047580.
- ↑ Barakat, Tahsin Stefan; Gribnau, Joost (2010). "X Chromosome Inactivation and Embryonic Stem Cells". The Cell Biology of Stem Cells. Landes Bioscience and Springer Science+Business Media. https://www.ncbi.nlm.nih.gov/books/NBK45037/.
- ↑ "X-inactivation informs variance-based testing for X-linked association of a quantitative trait". BMC Genomics 16 (1): 241. March 2015. doi:10.1186/s12864-015-1463-y. PMID 25880738.
- ↑ "Progression rate of myelopathy in X-linked adrenoleukodystrophy heterozygotes". Metabolic Brain Disease 30 (5): 1279–84. October 2015. doi:10.1007/s11011-015-9672-2. PMID 25920484.
- ↑ 23.0 23.1 23.2 "Genetic control of X inactivation and processes leading to X-inactivation skewing". American Journal of Human Genetics 58 (6): 1101–8. June 1996. PMID 8651285.
- ↑ 24.0 24.1 "Hemophagocytic lymphohistiocytosis in a female patient due to a heterozygous XIAP mutation and skewed X chromosome inactivation". Pediatric Blood & Cancer 62 (7): 1288–90. July 2015. doi:10.1002/pbc.25483. PMID 25801017.
- ↑ "Menkes disease with discordant phenotype in female monozygotic twins". American Journal of Medical Genetics. Part A 167A (11): 2826–9. November 2015. doi:10.1002/ajmg.a.37276. PMID 26239182.
- ↑ "Genetic characterization in symptomatic female DMD carriers: lack of relationship between X-inactivation, transcriptional DMD allele balancing and phenotype". BMC Medical Genetics 13: 73. August 2012. doi:10.1186/1471-2350-13-73. PMID 22894145.
- ↑ 27.0 27.1 "Long-range cis effects of ectopic X-inactivation centres on a mouse autosome". Nature 386 (6622): 275–9. March 1997. doi:10.1038/386275a0. PMID 9069285. Bibcode: 1997Natur.386..275L.
- ↑ 28.0 28.1 "Tsix, a gene antisense to Xist at the X-inactivation centre". Nature Genetics 21 (4): 400–4. April 1999. doi:10.1038/7734. PMID 10192391.
- ↑ 29.0 29.1 Mercer, T.R., Dinger, M.E., Mattick, J.S., (2009). Long non-coding RNAs: insight into functions. Nature Reviews Genetics. (10) 155–159.
- ↑ "A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse". Development 136 (1): 139–46. January 2009. doi:10.1242/dev.026427. PMID 19036803.
- ↑ "Xist and the order of silencing" (Review Article). EMBO Reports 8 (1): 34–9. January 2007. doi:10.1038/sj.embor.7400871. PMID 17203100. "Figure 1 Xist RNA encompasses the X from which it is transcribed.".
- ↑ "Requirement for Xist in X chromosome inactivation". Nature 379 (6561): 131–7. 1996. doi:10.1038/379131a0. PMID 8538762. Bibcode: 1996Natur.379..131P.
- ↑ 33.0 33.1 "Xist has properties of the X-chromosome inactivation centre". Nature 386 (6622): 272–5. March 1997. doi:10.1038/386272a0. PMID 9069284. Bibcode: 1997Natur.386..272H.
- ↑ "Xist and the order of silencing" (Review Article). EMBO Reports 8 (1): 34–9. January 2007. doi:10.1038/sj.embor.7400871. PMID 17203100. "Table 1 Features of the inactive X territory". – Originated from;
"Silencing of the mammalian X chromosome". Annual Review of Genomics and Human Genetics 6: 69–92. 2005. doi:10.1146/annurev.genom.6.080604.162350. PMID 16124854.
"Chromatin remodeling in dosage compensation". Annual Review of Genetics 39: 615–51. 2005. doi:10.1146/annurev.genet.39.073003.094210. PMID 16285873. - ↑ "Polycomb Repressor Complex 2 in Genomic Instability and Cancer". Int J Mol Sci 18 (8): 1657. July 2017. doi:10.3390/ijms18081657. PMID 28758948.
- ↑ "Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals". Nature 393 (6685): 599–601. June 1998. doi:10.1038/31275. PMID 9634239. Bibcode: 1998Natur.393..599C.
- ↑ "Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos". Development 127 (11): 2283–9. June 2000. doi:10.1242/dev.127.11.2283. PMID 10804171. http://dev.biologists.org/cgi/reprint/127/11/2283.pdf.
- ↑ "A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis". Nature 163 (4148): 676–677. April 1949. doi:10.1038/163676a0. PMID 18120749. Bibcode: 1949Natur.163..676B.
- ↑ "Xist RNA and the mechanism of X chromosome inactivation". Annual Review of Genetics 36: 233–78. 2002. doi:10.1146/annurev.genet.36.042902.092433. PMID 12429693.
- ↑ 40.0 40.1 "X-inactivation profile reveals extensive variability in X-linked gene expression in females". Nature 434 (7031): 400–4. March 2005. doi:10.1038/nature03479. PMID 15772666. Bibcode: 2005Natur.434..400C.
- ↑ 41.0 41.1 41.2 "Site-specific silencing of regulatory elements as a mechanism of X inactivation". Cell 151 (5): 951–63. November 2012. doi:10.1016/j.cell.2012.10.037. PMID 23178118.
- ↑ 42.0 42.1 "Global survey of escape from X inactivation by RNA-sequencing in mouse". Genome Research 20 (5): 614–22. May 2010. doi:10.1101/gr.103200.109. PMID 20363980.
- ↑ "Turner syndrome: MedlinePlus Genetics" (in en). https://medlineplus.gov/genetics/condition/turner-syndrome/.
- ↑ "Escape from X inactivation in mice and humans". Genome Biology 11 (6): 213. June 2010. doi:10.1186/gb-2010-11-6-213. PMID 20573260.
- ↑ "Glucose-6-phosphate dehydrogenase mosaicism: utilization as a cell marker in the study of leiomyomas". Science 150 (3692): 67–9. October 1965. doi:10.1126/science.150.3692.67. PMID 5833538. Bibcode: 1965Sci...150...67L.
- ↑ "Many different tumor types have polyclonal tumor origin: evidence and implications". Mutation Research 659 (3): 232–47. 2008. doi:10.1016/j.mrrev.2008.05.004. PMID 18614394. https://zenodo.org/record/1259237.
- ↑ "X-linked clonality testing: interpretation and limitations". Blood 110 (5): 1411–9. September 2007. doi:10.1182/blood-2006-09-018655. PMID 17435115.
- ↑ "The leiomyomatous stroma in renal cell carcinomas is polyclonal and not part of the neoplastic process". Virchows Archiv 465 (1): 89–96. July 2014. doi:10.1007/s00428-014-1591-9. PMID 24838683.
- ↑ "Evaluation of clonal origin of malignant mesothelioma". Journal of Translational Medicine 12: 301. December 2014. doi:10.1186/s12967-014-0301-3. PMID 25471750.
- ↑ "Translating dosage compensation to trisomy 21". Nature 500 (7462): 296–300. August 2013. doi:10.1038/nature12394. PMID 23863942. Bibcode: 2013Natur.500..296J.
- ↑ "Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus". Experimental Cell Research 18 (2): 415–8. October 1959. doi:10.1016/0014-4827(59)90031-X. PMID 14428474.
- ↑ "Gene action in the X-chromosome of the mouse (Mus musculus L.)". Nature 190 (4773): 372–3. April 1961. doi:10.1038/190372a0. PMID 13764598. Bibcode: 1961Natur.190..372L.
- ↑ "Glucose-6-phosphate dehydrogenase deficiency: a historical perspective". Blood 111 (1): 16–24. January 2008. doi:10.1182/blood-2007-04-077412. PMID 18156501.
- ↑ "The normal human female as a mosaic of X-chromosome activity: studies using the gene for C-6-PD-deficiency as a marker". Proceedings of the National Academy of Sciences of the United States of America 48 (1): 9–16. January 1962. doi:10.1073/pnas.48.1.9. PMID 13868717. Bibcode: 1962PNAS...48....9B.
Further reading
- "X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny". Nature Reviews. Genetics 6 (5): 410–8. May 2005. doi:10.1038/nrg1604. PMID 15818384.
- "Regulation of X-chromosome inactivation in development in mice and humans" (Review Article). Microbiology and Molecular Biology Reviews 62 (2): 362–78. June 1998. doi:10.1128/MMBR.62.2.362-378.1998. PMID 9618446.
- "The Lyon and the LINE hypothesis". Seminars in Cell & Developmental Biology 14 (6): 313–8. December 2003. doi:10.1016/j.semcdb.2003.09.015. PMID 15015738.
- "Xist and the order of silencing" (Review Article). EMBO Reports 8 (1): 34–9. January 2007. doi:10.1038/sj.embor.7400871. PMID 17203100.
- "Xist localization and function: new insights from multiple levels". Genome Biology 16 (1): 166. August 2015. doi:10.1186/s13059-015-0733-y. PMID 26282267.
External links
- Kruszelnicki, Karl (2004-02-11). "Hybrid Auto-Immune Women 3". ABC Science. http://www.abc.net.au/science/k2/moments/s1002754.htm.
 |