Biology:Sequential model
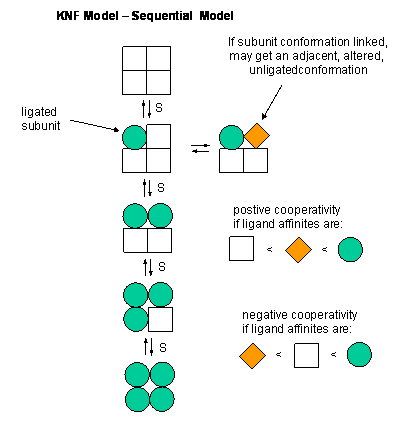
The sequential model (also known as the KNF model) is a theory that describes cooperativity of protein subunits.[1] It postulates that a protein's conformation changes with each binding of a ligand, thus sequentially changing its affinity for the ligand at neighboring binding sites. It gives one explanation for cooperative binding.
Overview
This model for allosteric regulation of enzymes suggests that the subunits of multimeric proteins have two conformational states.[1] The binding of the ligand causes conformational change in the other subunits of the multimeric protein. Although the subunits go through conformational changes independently (as opposed to in the MWC model), the switch of one subunit makes the other subunits more likely to change, by reducing the energy needed for subsequent subunits to undergo the same conformational change. In elaboration, the binding of a ligand to one subunit changes the protein's shape, thereby making it more thermodynamically favorable for the other subunits to switch conformation to the high affinity state. Ligand binding may also result in negative cooperativity, or a reduced affinity for the ligand at the next binding site, a feature that makes the KNF model distinct from the MWC model, which suggests only positive cooperativity.[2][3] It is named KNF after Koshland, Némethy and Filmer, who first suggested the model.[1]
History
A multimeric protein's affinity for a ligand changes upon binding to a ligand, a process known as cooperativity. This phenomenon was first discovered by Christian Bohr's analysis of hemoglobin, whose binding affinity for molecular oxygen increases as oxygen binds its subunits.[1] The concerted model (or MWC model or symmetry model) provides a theoretical basis for understanding this phenomenon. The model proposes that multimeric proteins exist in two separate states, T and R. Upon ligand binding, equilibrium between the two states shifts towards the R state, thought to result from protein conformation changes due to ligand binding. The model is useful in describing hemoglobin's sigmoidal binding curve.[4]
The KNF model (or induced fit model or sequential model) arose to address the possibility of differential binding states.[5] Developed by Koshland, Némethy and Filmer in 1966, the KNF model describes cooperativity as a sequential process, where ligand binding alters the conformation, and thus the affinity, of proximal subunits of the protein, resulting in several different conformations that have varying affinities for a given ligand. This model suggests that the MWC model oversimplifies cooperativity in that it does not account for conformational changes of individual binding sites, opting instead to suggest a single, whole-protein conformational change.[4]
Rules guiding the KNF model
The KNF model follows the structural theory of the induced fit model of substrate binding to an enzyme.[5] A slight change in the conformation of an enzyme improves its binding affinity to the transition state of the ligand, thus catalyzing a reaction. This follows the KNF model, which models cooperativity as the changing conformation of the ligand binding site upon ligand binding to another subunit.
Two essential assumptions guide the KNF model:[6]
- The protein exists in a single state of either low or high affinity for the ligand, when not bound to the ligand.......
- Upon ligation of a binding site, a conformational change is produced in that region of the protein. Changing this region of the protein may influence the conformation of nearby binding sites on the same protein, thus changing their affinity for the ligand. In negative cooperativity, affinity goes from high to low, while in positive cooperativity, affinity goes from low to high.
The KNF model characterizes enzymes that exhibit what was coined by Koshland and Hamadi in 2002 as i3 cooperativity.[2] This term is used merely to describe the structural nature of the sequential model, as the authors provide no other proposed descriptions or types of cooperativity.[7] These three properties are as follows:
- the nature of the subunits of the multimeric protein are such that they are identical to each other
- ligand binding induces a conformational change in the protein
- the conformational change is an intramolecular rearrangement within the protein
The i3 nature of a multimeric, cooperatively-acting protein is useful in standardizing the structural and physical basis of the sequential model.using model verification
Comparison to the MWC model
Structural differences
The primary differentiating feature between the MWC model and KNF model lies in the scale of conformational changes.[6] While both suggest that a protein's affinity for a given ligand changes upon binding of the ligand, the MWC model suggests that this occurs by a quaternary conformational change that involves the entire protein, moving from T state to favoring the R state. On the other hand, the KNF model suggests these conformational changes occur on the level of tertiary structure within the protein, as neighboring subunits change conformation with successive ligand binding.[8]
Unlike the MWC model, the KNF model offers the possibility of "negative cooperativity".[2][6] This term describes a reduction in the affinity of the other binding sites of a protein for a ligand after the binding of one or more of the ligand to its subunits. The MWC model only allows for positive cooperativity, where a single conformational switch from the T to R states results in an increase in affinity for the ligand at unligated binding sites. Ligand binding to the T state thus cannot increase the amount of the protein in the T, or low-affinity, state.
Negative cooperativity is observed in a number of biologically significant molecules, including tyrosyl-tRNA synthetase and glyceraldehyde-3-phosphate dehydrogenase.[3][6] In fact, in a systematic literature review performed in 2002 by Koshland and Hamadani, the same literature review that coined i3 cooperativity, negatively cooperating proteins are seen to compose slightly less than 50% of scientifically studied proteins that exhibit cooperativity, while positively cooperating proteins compose the other, slightly greater than 50%.[2]
Functional differences in hemoglobin
Hemoglobin, a tetrameric protein that transports four molecules of oxygen, is a highly biologically relevant protein that has been a subject of debate in allostery. It exhibits a sigmoidal binding curve, indicating cooperativity. While most scientific evidence points to concerted cooperativity,[9][10] research into the affinities of specific heme subunits for oxygen has revealed that under certain physiological conditions, the subunits may display properties of sequential allostery.[11] Nuclear magnetic resonance (NMR) studies show that in the presence of phosphate, deoxygenated human adult hemoglobin's alpha heme subunits display increased affinity for molecular oxygen, when compared to beta subunits. The results suggest either a modified concerted model, in which alpha subunits have a greater affinity for oxygen in the quaternary low-affinity T state, or a sequential model, in which phosphate binding creates a partially oligomerized state that stabilizes a low affinity form of the beta subunits, distinct from a T or R state.[11] Thus, depending on physiological conditions, a combination of the MWC and KNF models appears to most comprehensively describe hemoglobin's binding characteristics.[9]
References
- ↑ 1.0 1.1 1.2 Koshland, D.E., Némethy, G. and Filmer, D. (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385. DOI: 10.1021/bi00865a047
- ↑ 2.0 2.1 2.2 2.3 Koshland, Daniel E.; Hamadani, Kambiz (2002-12-06). "Proteomics and Models for Enzyme Cooperativity" (in en). Journal of Biological Chemistry 277 (49): 46841–46844. doi:10.1074/jbc.R200014200. ISSN 0021-9258. PMID 12189158.
- ↑ 3.0 3.1 Henis, Y I; Levitzki, A (1980-09-01). "Mechanism of negative cooperativity in glyceraldehyde-3-phosphate dehydrogenase deduced from ligand competition experiments.". Proceedings of the National Academy of Sciences of the United States of America 77 (9): 5055–5059. doi:10.1073/pnas.77.9.5055. ISSN 0027-8424. PMID 6933545. Bibcode: 1980PNAS...77.5055H.
- ↑ 4.0 4.1 Marzen, Sarah; Garcia, Hernan G.; Phillips, Rob (2013-05-13). "Statistical mechanics of Monod-Wyman-Changeux (MWC) models". Journal of Molecular Biology 425 (9): 1433–1460. doi:10.1016/j.jmb.2013.03.013. ISSN 1089-8638. PMID 23499654.
- ↑ 5.0 5.1 "Model Binding Systems" (in en-US). Biology LibreTexts. 2013-11-21. http://bio.libretexts.org/Core/Biochemistry/Binding/MODEL_BINDING_SYSTEMS#Free_Energy_and_Cooperativity.
- ↑ 6.0 6.1 6.2 6.3 Alan, Fersht (1999). Structure and mechanism in protein science : a guide to enzyme catalysis and protein folding. Freeman. ISBN 9780716732686. OCLC 837581840.
- ↑ Purich, Daniel L. (2010-06-16) (in en). Enzyme Kinetics: Catalysis and Control: A Reference of Theory and Best-Practice Methods. Elsevier. ISBN 9780123809254. https://books.google.com/books?id=SkSQNNACcrYC&q=i3+cooperativity&pg=PA687.
- ↑ Ronda, Luca; Bruno, Stefano; Bettati, Stefano (2013-09-01). "Tertiary and quaternary effects in the allosteric regulation of animal hemoglobins". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. Oxygen Binding and Sensing Proteins 1834 (9): 1860–1872. doi:10.1016/j.bbapap.2013.03.013. PMID 23523886.
- ↑ 9.0 9.1 Cui, Qiang; Karplus, Martin (2017-03-25). "Allostery and cooperativity revisited". Protein Science 17 (8): 1295–1307. doi:10.1110/ps.03259908. ISSN 0961-8368. PMID 18560010.
- ↑ Berg, Jeremy M.; Tymoczko, John L.; Stryer, Lubert (2002-01-01) (in en). Hemoglobin Transports Oxygen Efficiently by Binding Oxygen Cooperatively. https://www.ncbi.nlm.nih.gov/books/NBK22596/.
- ↑ 11.0 11.1 Lindstrom, Ted (1972). "Functional nonequivalence of alpha and beta hemes in human adult hemoglobin". Proceedings of the National Academy of Sciences 69 (7): 1707–1710. doi:10.1073/pnas.69.7.1707. PMID 4505648. Bibcode: 1972PNAS...69.1707L.
 |