Biology:Photoactive yellow protein
| PYP | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PYP | ||||||||
| Pfam | PF00989 | ||||||||
| InterPro | IPR012130 | ||||||||
| SMART | SM00091 | ||||||||
| PROSITE | PS50112 | ||||||||
| SCOP2 | 55786 / SCOPe / SUPFAM | ||||||||
| CDD | cd00130 | ||||||||
| |||||||||
In molecular biology, the PYP domain (photoactive yellow protein) is a p-coumaric acid-binding protein domain. They are present in various proteins in bacteria.
PYP is a highly soluble globular protein with an alpha/beta fold structure. It is a member of the PAS domain superfamily, which also contains a variety of other kinds of photosensory proteins.
PYP was first discovered in 1985.[1]
A recently (2016) developed chemogenetic system named FAST (Fluorescence-Activating and absorption Shifting Tag) was engineered from PYP to specifically and reversibly bind a series of hydroxybenzylidene rhodanine (HBR) derivatives for their fluorogenic properties. Upon interaction with FAST, the fluorogen is locked into a fluorescent conformation unlike when in solution. This new protein labelling system is used in a variety of microscopy and cytometry setups.[2]
p-Coumaric acid
p-Coumaric acid is a cofactor of Photoactive yellow protein|photoactive yellow proteins.[3] Adducts of p-coumaric acid bound to PYP form crystals that diffract well for x-ray crystallography experiments. These structural studies have provided insight into photosensitive proteins, e.g. the role of hydrogen bonding, molecular isomerization and photoactivity.[4][5][6][7]
Photochemical transitions
It was originally believed that due to light emissions resembling that of retinal bound rhodopsin, the photosensor molecule bound to PYP should resemble the structure of retinal bound rhodopsin, the photosensor molecule bound to PYP should resemble the structure of retinal.[8] Scientists were therefore amazed when the PYP Cys 69 was bound by a thiol ester linkage as the light sensitive prosthetic group p-coumaric acid.[3] During the photoreactive mechanism:[3][8]
- Light absorption yields the native protein to absorb a maximum wavelength of 446 nm, ε = 45500 M−1 cm−1.
- Within a nanosecond the absorbed maximum wavelength is shifted to 465 nm.
- Then on a sub-millisecond timescale is excited to a 355 nm state.
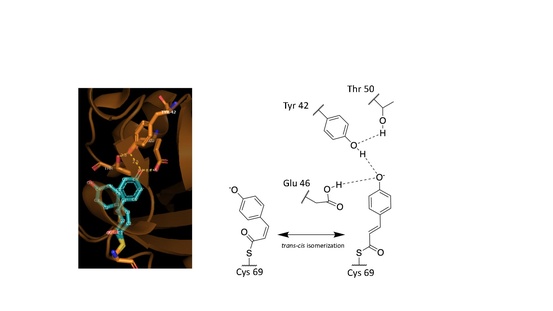
These observed phenomena are due to the trans–cis isomerization of the vinyl trans double bond in the p-coumaric acid.[3][7][6] Scientists noted by observing the crystal structure of p-coumaric acid bound by PYP that the hydroxyl group connected to the C4 carbon of the phenyl ring appeared to be deprotonated – effectively a phenolate functional group.[7][10] This was due to abnormally short hydrogen bonding lengths observed in the protein crystal structure.[9]
Role of hydrogen bonding
Hydrogen bonds in proteins such as PYP take part in interrelated networks, where at the center of p-coumaric acid's phenolate O4 atom, there is an oxyanion hole that is crucial for photosensory function.[6][11][12] Oxyanion holes exist in enzymes to stabilize transitions states of reaction intermediates, thus stabilizing the trans–cis isomerization of p-coumaric acid.[5][13] During the transition state it is believed that the p-coumaric acid phenolate O4 takes part in a hydrogen bond network between Glu46, Tyr42 and Thr50 of PYP.[13][5] These interactions are apart from the thiol ester linkage to Cys 69 keeping p-coumaric acid in the ligand binding site.[3] Upon transitioning to the cis-isomeric form of p-coumaric acid the favorable hydrogen bonds are no longer in close interaction.
References
- ↑ "Isolation and characterization of soluble cytochromes, ferredoxins and other chromophoric proteins from the halophilic phototrophic bacterium Ectothiorhodospira halophila". Biochimica et Biophysica Acta (BBA) - Bioenergetics 806 (1): 175–83. January 1985. doi:10.1016/0005-2728(85)90094-5. PMID 2981543.
- ↑ "Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo". Proceedings of the National Academy of Sciences of the United States of America 113 (3): 497–502. January 2016. doi:10.1073/pnas.1513094113. PMID 26711992.
- ↑ 3.0 3.1 3.2 3.3 3.4 "Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry". Biochemistry 33 (47): 13959–62. November 1994. doi:10.1021/bi00251a001. PMID 7947803. http://dare.uva.nl/personal/pure/en/publications/pcoumaric-acid-a-new-photoactive-chromophore-of-a-yellow-photoreceptor-protein-with-rhodopsinlike-characteristics(6bd029db-bd7c-439b-af3a-46357240b242).html.
- ↑ "PDB101: Molecule of the Month: Photoactive Yellow Protein". http://pdb101.rcsb.org/motm/207.
- ↑ 5.0 5.1 5.2 "Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein". Journal of the American Chemical Society 140 (31): 9827–9843. August 2018. doi:10.1021/jacs.8b01596. PMID 29990421.
- ↑ 6.0 6.1 6.2 "Key issues in the photochemistry and signalling-state formation of photosensor proteins". Journal of Photochemistry and Photobiology B: Biology 54 (2–3): 94–102. February 2000. doi:10.1016/S1011-1344(00)00004-X. PMID 10836537.
- ↑ 7.0 7.1 7.2 "Impact of Photon Absorption on the Electronic Properties of p -Coumaric Acid Derivatives of the Photoactive Yellow Protein Chromophore". The Journal of Physical Chemistry B 108 (16): 5138–5148. 2004-01-30. doi:10.1021/jp037469b.
- ↑ 8.0 8.1 "Properties of a water-soluble, yellow protein isolated from a halophilic phototrophic bacterium that has photochemical activity analogous to sensory rhodopsin". Biochemistry 26 (2): 418–23. January 1987. doi:10.1021/bi00376a012. PMID 3828315.
- ↑ 9.0 9.1 "Structure of a protein photocycle intermediate by millisecond time-resolved crystallography". Science 275 (5305): 1471–5. March 1997. doi:10.1126/science.275.5305.1471. PMID 9045611.
- ↑ "Low-barrier hydrogen bond in photoactive yellow protein". Proceedings of the National Academy of Sciences of the United States of America 106 (2): 440–4. January 2009. doi:10.1073/pnas.0811882106. PMID 19122140. Bibcode: 2009PNAS..106..440Y.
- ↑ "1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore". Biochemistry 34 (19): 6278–87. May 1995. doi:10.1021/bi00019a004. PMID 7756254.
- ↑ "On the involvement of single-bond rotation in the primary photochemistry of photoactive yellow protein". Biophysical Journal 101 (5): 1184–92. September 2011. doi:10.1016/j.bpj.2011.06.065. PMID 21889456. Bibcode: 2011BpJ...101.1184S.
- ↑ 13.0 13.1 "Hydrogen Bonds: Simple after All?". Biochemistry 57 (24): 3338–3352. June 2018. doi:10.1021/acs.biochem.8b00217. PMID 29678112.
Further reading
- "Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily". Photochemistry and Photobiology 83 (1): 40–9. 2007. doi:10.1562/2006-02-28-IR-827. PMID 16939366.
External links
- Overview of all the structural information available in the PDB for UniProt: P16113 (Photoactive yellow protein) at the PDBe-KB.
 |

