Biology:MIPOL1
MIPOL1 (Mirror Image Polydactyly 1), also known as CCDC193 (Coiled-coil domain containing 193), is a protein that in humans is encoded by the MIPOL1 gene.[1][2] Mutation of this gene is associated with mirror-image polydactyly (also known as Laurin-Sandrow syndrome.[3]) in humans, which is a rare genetic condition characterized by mirror-image duplication of digits.[4]
Gene
MIPOL1 is also known as CCDC193 (Coiled-coil domain containing 193).
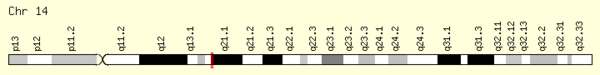
Locus
The MIPOL1 gene is located at 14q13.3-q21.1 on the plus strand, spanning base pairs 37,197,888 to 37,579,207 (in the human GRCh38 primary assembly, length: 381,320 base pairs), consisting of 15 exons and 11 introns. Some notable genes in its neighborhood include SLC25A21 (mutation of this gene causes synpolydactyly[6]) and FOXA1.
mRNA
MIPOL1 has at least 15 known splice isoforms produced by alternative splicing.[7]
Protein
Properties
The unmodified MIPOL1 protein isoform 1 in humans has an isoelectric point of 5.6 and molecular weight 51.5 kDa.[8] Relative to other human proteins, MIPOL1 consists of unusually low amounts of Proline and Glycine and higher amounts of Glutamic acid and Glutamine.[9]
Isoforms
There are at least three known isoforms of this protein in humans produced by alternative splicing: isoform 1, of length 442 amino acids, isoform 2 of length 261 amino acids and isoform 3 of length 169 amino acids.[1]
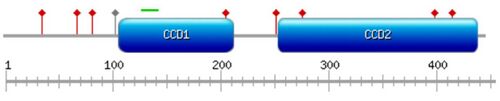
Domains and motifs
MIPOL1 contains two coiled-coil domains in its C-terminus at positions 107 – 212 and 253 – 435[1] (shown in Fig.1). A bipartite nuclear localization signal is predicted at position 128 – 143.[12]

Post-translational modifications
The following post-translational modifications are predicted using bioinformatics tools for MIPOL1.[13] Multiple phosphorylation sites are predicted for this protein, that are conserved in close orthologs, including a Casein kinase 1 (CK1) site, three Casein kinase 2 (CK2) sites, and three NEK2 sites.[14]
| Post-translational modification | Amino acid site | Prediction tool |
|---|---|---|
| Phosphorylation | Ser (37, 42, 69, 75, 105, 126, 205, 275, 344, 350, 364, 412), Thr (80, 251, 259, 338, 365, 396, 435, 437, 440), Tyr (77, 83) | MyHits,[12] NetPhos[15] |
| O-linked glycosylation | Ser (34, 105, 294, 344, 412), Thr (155, 293) | NetOGlyc[16] |
| O-GlcNAcylation | Thr (104) | YinOYang[17] |
| Glycation | Lys (6, 41, 133, 207, 347, 421) | NetGlycate[18] |
| SUMOylation | Lys (136, 147) | GPS-SUMO[19] |
| Ubiquitination | Lys (22, 47, 133, 162, 314, 418) | BDM-PUB[20] |

Structure
The exact structure of the MIPOL1 has not yet been characterized. Homology-based and de novo predictions of its tertiary structure suggest that it may consist of inter-twined alpha helices, forming coiled-coil domains (see Fig.4.).[22][21]
Sub-cellular localization
Immunofluorescence imaging in the human U2OS cell line (bone Osteosarcoma epithelial cells) shows localization in the cytosol.[23] Immunohistochemistry imaging of human prostate tissue also suggests cytosolic localization.[24] A bipartite nuclear localization signal is predicted at position 128 – 143, which is highly conserved in mammalian orthologs (see Fig.2.), indicating possible localization in the nucleus.[12]
Gene regulation
The predicted promoter sequence for this gene spans from base pair 37196852 to 37198126 (1,275 bp) and has multiple predicted binding sites for transcription factors such as GATA binding factors, SMAD3, TP63 and NRF1.[25]
Gene Expression
MIPOL1 is ubiquitously expressed at low levels in humans, with highest expression in the prostate.[1]
Transcript regulation
The RNA secondary structure is stabilized by multiple stem loops that have been predicted (using bioinformatics tools[26]), and conserved across closely related species. Multiple binding targets are found for microRNAs such as MIR3163 and MIR190a, that could silence these regions on the mRNA and inhibit translation.[27]
Clinical significance
The MIPOL1 gene is an autosomal dominant gene.[28] It is one of six genes in humans causing non-syndromic polydactyly (i.e. polydactyly occurring as a separate event with no other associated anomalies).[29] Mutation of this gene is associated with mirror-image polydactyly (also known as Laurin-Sandrow syndrome[28]) in humans, which is a rare genetic condition characterized by mirror-image duplication of digits in hands and feet.[4]
This gene has also been associated with central nervous system development, and the loss of this gene can cause craniofacial defects and agenesis of the corpus callosum.[30]
The gene is shown to function as a tumor suppressor in nasopharyngeal carcinoma (NPC), through the up-regulation of the p21 (WAF1/CIP1) and p27 (proteins that are both cyclin-dependent kinases that are linked with tumor suppression via cell cycle arrest) pathways.[31] Another study investigating the role of MIPOL1 gene in cancer progression reported that MIPOL1 was downregulated in NPC tumor tissues, and that artificially re-expressing the gene caused tumor suppression by down-regulating angiogenic factors and reducing the phosphorylation of metastasis associated proteins like AKT, p65 and FAK14.[32] MIPOL1 interacts with another well-known tumor-suppressing gene, RhoB and this interaction was confirmed to enhance RhoB activity.
In a study of pediatric high grade glioma (pHGG), MIPOL1 gene was found to be down-regulated 2.4-fold in the high vascularity tumors[33]
The protein is known to interact with Replicase polyprotein 1ab in SARS-CoV2, which is a protein involved in the transcription and replication of viral RNAs.[34]
Interacting proteins
This protein is known to interact with multiple human proteins, verified via two-hybrid screening. A few notable examples include:
LATS2: Negatively regulates YAP1 in the Hippo signaling pathway that plays a pivotal role in organ size control and tumor suppression by restricting cell proliferation and promoting apoptosis.[35]
ZGPAT (Zinc finger CCCH-type with G patch domain-containing protein): A transcription repressor that negatively regulates expression of EGFR, a gene involved in cell proliferation, survival and migration, suggesting that it may act as a tumor suppressor.[36]
RCOR3 (REST Corepressor 3): A protein that may act as a component of a co-repressor complex that represses transcription[37]
It also interacts with viral proteins such as:
Replicase polyprotein 1ab (SARS-CoV2): A multifunctional protein involved in the transcription and replication of viral RNAs.[34]
Protein E7 (Human Papillomavirus): Plays a role in viral genome replication by driving entry of quiescent cells into the cell cycle.[38]
Origin and evolution
The earliest known ortholog of this protein appeared around 948 million years ago in Trichoplax adhaerens in phylum Placozoa in kingdom Animalia. The next most distant orthologs appear in phylum Cnidaria, around 824 million years ago.
Sequence Homology
The MIPOL1 protein has no known paralogs in humans and other species for which orthologs have been found, therefore, it is the only member of its gene family.
There are more than 300 known orthologs of the MIPOL1 protein in Animalia, ranging from primates to corals and sea anemones in phylum Cnidaria.[39] Orthologs of the protein were found in species as distant as Trichoplax adhaerens, a simple primitive invertebrate species. Table 2 shows a sample of the ortholog space.
Closely related orthologs are found in chordates such as mammals, reptiles, birds and amphibians, with sequence similarities greater than 70%. Sequence lengths of orthologs were similar to the human MIPOL1 protein, with no significant gene duplication observed.
Organisms with sequence similarities in the 55-70% range (moderately related orthologs) were found in bony fish, cartilaginous fish and coelacanths. Sequence length is generally longer in these species, with a longer amino acid sequence in the N-terminus (alignment with human protein occurs around amino acid 100).
Distantly related orthologs with similarities less than 50% (around 30 – 40%) are found in hemichordates, echinoderms, arthropods, molluscs, cnidaria and placozoa. Multiple sequence alignment with distant orthologs indicates poor alignment in the N-terminus of the protein.
Two COG (Clusters of Orthologous Groups of proteins) domains were found in this protein (see Fig.3): COG1196 at position 106 - 340 (Chromosome segregation ATPase[40]) and COG4372 at 259 - 431 (uncharacterized conserved protein containing a DUF3084 domain[41])[42]
| Species | Organism Common Name | NCBI Accession | Date of Divergence from humans (MYA)[43] | Sequence length (AAs) | Sequence Identity to human protein (%) |
|---|---|---|---|---|---|
| Homo sapiens | Human | NP_001182226.1 | 0 | 442 | 100 |
| Macaca fascicularis | Crab-eating macaque | XP_005561170.1 | 29.44 | 442 | 94.3 |
| Lipotes vexillifer | Yangtze River dolphin | XP_007465863.1 | 96 | 442 | 85.5 |
| Manis javanica | Malayan pangolin | XP_017524645.1 | 96 | 407 | 77.6 |
| Myotis brandtii | Brandt's bat | XP_005865039.1 | 96 | 463 | 74.6 |
| Chelonia mydas | Green sea turtle | XP_007065307.1 | 312 | 440 | 67.9 |
| Notechis scutatus | Mainland tiger snake | XP_026520263.1 | 312 | 429 | 54.0 |
| Gallus gallus | Chicken | XP_004941823.1 | 312 | 429 | 55.9 |
| Rhinatrema bivittatum | Two-lined caecilian | XP_029454767.1 | 351.8 | 443 | 59.3 |
| Latimeria chalumnae | West Indian Ocean coelacanth | XP_005989227.1 | 413 | 532 | 51.9 |
| Danio rerio | Zebrafish | XP_021322786.1 | 435 | 381 | 38.8 |
| Oryzias melastigma | Indian medaka | XP_024129547.1 | 435 | 404 | 30.9 |
| Amblyraja radiata | Thorny skate | XP_032883021.1 | 473 | 497 | 45.8 |
| Saccoglossus kowalevskii | Acorn worm | XP_006815617.1 | 684 | 666 | 23.3 |
| Asterias rubens | Common starfish | XP_033636481.1 | 684 | 646 | 22.2 |
| Limulus polyphemus | Atlantic horseshoe crab | XP_022249371.1 | 797 | 579 | 19.2 |
| Pecten maximus | Great scallop | XP_033734074.1 | 797 | 598 | 22.5 |
| Exaiptasia pallida | Brown anemone | KXJ12639.1 | 824 | 468 | 27.5 |
| Orbicella faveolata | Mountainous star coral | XP_020610356.1 | 824 | 453 | 27.5 |
| Trichoplax adhaerens | Trichoplax | XP_002117892.1 | 948 | 553 | 22.9 |
Phylogenetics
Using a linear regression analysis on a plot of corrected percent divergence (amino acid changes per 100 amino acids) as a function of date of divergence from humans for different MIPOL1 orthologs (see Fig.5), it is estimated that a 1% change in amino acids in the MIPOL1 protein takes 5.68 million years. MIPOL1 protein is evolving at a moderate rate relative to fast evolving protein such as fibrinogen alpha, and slow evolving proteins such as cytochrome C.
References
- ↑ 1.0 1.1 1.2 1.3 NCBI Gene Mirror-Image Polydactyly 1. Retrieved 27 July 2020.
- ↑ "MIPOL1 - Mirror-image polydactyly gene 1 protein - Homo sapiens (Human) - MIPOL1 gene & protein". Uniprot. https://www.uniprot.org/uniprot/Q8TD10.
- ↑ OMIM Entry on Laurin-Sandrow Syndrome (Mirror-Image Polydactyly). Retrieved 27 July 2020.
- ↑ 4.0 4.1 Kondoh S, Sugawara H, Harada N, et al. A novel gene is disrupted at a 14q13 breakpoint of t(2;14) in a patient with mirror-image polydactyly of hands and feet. J Hum Genet. 2002;47(3):136-139. doi:10.1007/s1003802000 15
- ↑ Hubbard T, Barker D, Birney E, et al. "The Ensembl genome database project." Nucleic Acids Res. 2002;30(1):38-41. doi:10.1093/nar/30.1.38
- ↑ Meyertholen K, Ravnan JB, Matalon R. Identification of a Novel 14q13.3 Deletion Involving the SLC25A21 Gene Associated with Familial Synpolydactyly. Mol Syndromol. 2012;3(1):25-29. doi:10.1159/000339177
- ↑ Ensembl Entry on MIPOL1. Retrieved 30 July 2020
- ↑ ExPASy Compute pI/MW tool. Retrieved 27 July 2020.
- ↑ SAPS (Statistical Analysis of Protein Sequences) - Compositional Analysis. Retrieved 27 July 2020
- ↑ Prosite MyDomains tool. Retrieved 27 July 2020.
- ↑ Nardozzi, J. D., Lott, K., & Cingolani, G. (2010). Phosphorylation meets nuclear import: a review. Cell communication and signaling : CCS, 8, 32. https://doi.org/10.1186/1478-811X-8-32
- ↑ 12.0 12.1 12.2 Swiss Institute of Bioinformatics MyHits Motif Scan. Retrieved 27 July 2020.
- ↑ Swiss Institute of Bioinformatics ExPASy Portal. Retrieved 27 July 2020
- ↑ Eukaryotic Linear Motif resource. Retrieved 27 July 2020.
- ↑ NetPhos 3.1. Retrieved 27 July 2020.
- ↑ NetOGlyc. Retrieved 27 July 2020
- ↑ YinOYang. Retrieved 27 July 2020
- ↑ NetGlycate. Retrieved 27 July 2020
- ↑ GPS-SUMO. Retrieved 27 July 2020
- ↑ BDM-PUB. Retrieved 27 July 2020
- ↑ 21.0 21.1 I-TASSER Server for protein structure and function prediction. Retrieved 20 July 2020
- ↑ Coiled-coil prediction. Retrieved 27 July 2020
- ↑ Thermo Fisher Scientific anti-MIPOL1 Antibody produced in rabbit (PA5-65599). Retrieved 27 July 2020.
- ↑ Sigma Aldrich anti-MIPOL1 polyclonal antibody produced in rabbit (HPA002893). Retrieved 27 July 2020.
- ↑ ElDorado Genomatix regulatory analysis tools. Retrieved 13 July 2020.
- ↑ RNA secondary structure prediction. Retrieved 13 July 2020.
- ↑ miRDB microRNA database. Retrieved 2 July 2020.
- ↑ 28.0 28.1 OMIM Entry on Laurin-Sandrow Syndrome (Mirror-Image Polydactyly). Retrieved 27 July 2020
- ↑ Umair M, Ahmad F, Bilal M, Ahmad W, Alfadhel M. Clinical Genetics of Polydactyly: An Updated Review. Front Genet. 2018;9:447. Published 2018 Nov 6. doi:10.3389/fgene.2018.00447
- ↑ Shaffer JR, Orlova E, Lee MK, et al. Genome-Wide Association Study Reveals Multiple Loci Influencing Normal Human Facial Morphology. PLoS Genet. 2016;12(8):e1006149. Published 2016 Aug 25. doi:10.1371/journal.pgen.1006149
- ↑ Cheung AK, Lung HL, Ko JM, et al. Chromosome 14 transfer and functional studies identify a candidate tumor suppressor gene, mirror image polydactyly 1, in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2009;106(34):14478-14483. doi:10.1073/pnas.0900198106
- ↑ Leong MML, Cheung AKL, Kwok TCT, Lung ML. Functional characterization of a candidate tumor suppressor gene, Mirror Image Polydactyly 1, in nasopharyngeal carcinoma. Int J Cancer. 2020;146(10):2891-2900. doi:10.1002/ijc.32732
- ↑ Smith SJ, Tilly H, Ward JH, et al. CD105 (Endoglin) exerts prognostic effects via its role in the microvascular niche of paediatric high grade glioma. Acta Neuropathol. 2012;124(1):99-110. doi:10.1007/s00401-012-0952-1
- ↑ 34.0 34.1 UniProt entry on Replicase 1ab. Retrieved 27 July 2020.
- ↑ UniProt entry on LATS2. Retrieved 27 July 2020.
- ↑ UniProt entry on ZGPAT. Retrieved 27 July 2020.
- ↑ UniProt entry on RCOR3. Retrieved 27 July 2020.
- ↑ UniProt entry on protein E7. Retrieved 27 July 2020.
- ↑ NCBI entry on MIPOL1 orthologs. Retrieved 30 June 2020
- ↑ NCBI Conserved Protein Domain Family entry on COG1196. Retrieved 10 June 2020.
- ↑ NCBI Conserved Protein Domain Family entry on COG4372. Retrieved 10 June 2020
- ↑ Tatusov, R. L., Galperin, M. Y., Natale, D. A., & Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic acids research, 28(1), 33–36. https://doi.org/10.1093/nar/28.1.33
- ↑ Time tree: Approximate divergence between two taxa. Retrieved 30 June 2020.
 |




