Biology:L-Tryptophan decarboxylase
| alcohol dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
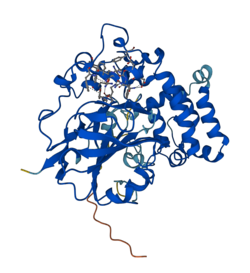 Crystal structure prediction for L--tryptophan decarboxylase .[1] | |||||||||
| Identifiers | |||||||||
| EC number | 4.1.1.105 | ||||||||
| Alt. names | aldehyde reductase | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
L-Tryptophan decarboxylase (EC 4.1.1.105) is an enzyme distinguished by the substrate L-tryptophan.[2][3]
This enzyme catalyzes the reaction of L-tryptophan to tryptamine and carbon dioxide.[2][4] The enzymatic reaction namely takes place in the species Psilocybe cubensis, where a decarboxylase, kinase, and methyltransferase work together to synthesize psilocybin.[5][6]
Classification
The enzyme commission number for L-tryptophan decarboxylase is EC 4.1.1.105.[2] Other common names include psilocybin biosynthesis decarboxylase and psiD.[4] The first digit in the enzyme number is representative of the class of enzymes known as lyases, which catalyze elimination reactions.[2][4] The second and third digits are representative of the subclass of lyases known as decarboxylases that cleave carbon-carbon bonds.[2][4] The last digit is representative of the enzyme’s specific substrate, L-tryptophan.[2]
This enzyme is a part of the PLP-independent phosphatidylserine decarboxylase family and most compared to hypothetical proteins of other basidiomycetes fungi.[5] These include Fibulorhizoctonia sp with 60% identical amino acids and Moniliophthora roreri with 52% identical amino acids.[5] A similar enzyme that is not related to L-tryptophan decarboxylase is called aromatic-L-amino-acid decarboxylase with an enzyme number of EC 4.1.1.28.[2]
Reaction pathway
The first step in the reaction is the substrate binding of L-tryptophan, which reacts with a coenzyme hydrogen.[2] The decarboxylase enzyme is able to transform L-tryptophan to tryptamine in the second step by cleaving off two oxygens and a carbon to form tryptamine and carbon dioxide as the products.[2] Tryptophan contains an α-amino group, an α-carboxylic acid group, and a side chain indole, which makes the molecule polar, while tryptamines have an indole ring structure, a fused double ring consisting of a pyrrole ring, and a benzene ring, which is joined to an amino group by two carbon side chains.[7]
This is the chemical reaction that takes place:
Organisms in which L-tryptophan decarboxylase is found
L-Tryptophan decarboxylase has been characterized in bacteria, plants, and fungi.[2] Fungi that produce psilocybin and psilocin express incredible diversity, as they are a part of at least eight genera with hundreds of species belonging to them.[3] The specific reaction pathway for L-tryptophan decarboxylase has been described in twelve species, including Psilocybe cubensis, commonly known as magic mushrooms.[2] All fungi in the genus Psilocybe have a well-defined, umbrella-like cap with gills underneath and a stipe.[3] Other main characteristics of Psilocybe species include purple-brown reproductive spores, the presence of an annulus, and blue bruising with contact.[3] All Psilocybe species are described to feed on microscopic detritus and are found on a variety of surfaces, such as herbivore dung, grasses, roots, wood, and soil.[3] Humans have a documented history of ingesting this psilocybin producing fungi.[3] There are 57 species that are found in Mexico, and out of these there have been reports of 35 species and nine varieties being used by ethnic groups.[3]
Function
In Psilocybe cubensis, L-tryptophan decarboxylase has been described with two other enzymes to biosynthesize psilocybin in a one pot reaction.[4][5] These other two enzymes in this process are psiK, an enzyme that catalyzes the phosphotransfer step, and psiM, an enzyme that catalyzes the iterative N-methyl transfer step.[4] The biosynthesis of psilocybin takes place as follows:
L-Tryptophan is decarboxylated to 4-hydroxytryptamine by psiD → psiK phosphorylates 4-hydroxytryptamine to create norbaeocystin → psiM then processively N,N-dimethylates the compound to yield psilocybin.[4]
Serotonin 2A receptors (5-HT2ARs) stimulation by the active metabolite, psilocin, disrupts serotonergic neurotransmission and produces the characteristic psychedelic effects of this species of fungus.[6] The product formed by L-tryptophan decarboxylase, tryptamine, is relevant to humans because the mammalian brain contains very low concentrations of tryptamine; and serotonin is a tryptamine natural derivative involved in regulating central nervous system processes like sleep, cognition, memory, temperature regulation and behavior.[7] In contrast, the naturally occurring derivatives of tryptamine are found in magic mushrooms (Psilocybe cubensis).[7]
Structure
L-Tryptophan decarboxylase is 439 amino acid residues long in its native form and a calculated pI 5.3.[4] The crystal structure of L-tryptophan decarboxylase has been modeled and predicted by AlphaFold with an average confidence of 91.17% and SWISS-MODEL with an average confidence of 25.37% as an oligo-state monomer, but the crystal structure remains to be described.[1][8]
Active sites
4-Hydroxy-L-tryptophan is accepted as a substrate by the enzyme in addition to L-tryptophan.[4][5] This subsequent pathway is suggested to yield 4-Hydroxytryptamine instead of tryptamine.[4] Both of these compounds can be used in the biosynthesis of psilocybin.[4] The enzyme is distinct from other fungal and plant aromatic amino acid decarboxylases because it belongs to a class that L-tryptophan has not previously been described as a substrate for.[4] Currently, the active sites for L-tryptophan decarboxylase remain to be described.
Evolution
Due to the multi-step process of psilocybin biosynthesis and its restricted phylogenetic distribution, the pathway involving L-tryptophan decarboxylase has been suggested to evolve via horizontal gene cluster transfer.[9][10] The phylogenies of the genes involved in psilocybin biosynthesis (including L-tryptophan decarboxylase) suggests that the process first evolved in wood-decaying fungi, and then evolved in dung-decaying fungi through vertical and horizontal gene transfer due to shared environmental pressures.[9][10] Neurological effects from psilocybin in wood and dung decaying fungi posit that psilocybin could be an ecological modulator acting on insect behavior.[9] This would advantage the fungi by disrupting and inhibiting the behavior of competitors, such as termites and fruit fly larvae, for wood and dung resources.[9][11] Invertebrate competitors that would be especially impacted by this are social insects, where neurotransmitter mimics would disrupt coordination.[12]
References
- ↑ 1.0 1.1 "L-tryptophan decarboxylase". AlphaFold Protein Structure Database. https://alphafold.ebi.ac.uk/entry/P0DPA6.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 "Information on EC 4.1.1.105 - L-tryptophan decarboxylase". BRENDA Enzyme Database. https://www.brenda-enzymes.org/enzyme.php?ecno=4.1.1.105.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 "Diversity, biology, and history of psilocybin-containing fungi: Suggestions for research and technological development". Fungal Biology 126 (4): 308–319. April 2022. doi:10.1016/j.funbio.2022.01.003. PMID 35314062.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 "Enzymatic Synthesis of Psilocybin". Angewandte Chemie 56 (40): 12352–12355. September 2017. doi:10.1002/anie.201705489. PMID 28763571.
- ↑ 5.0 5.1 5.2 5.3 5.4 "Biocatalytic Production of Psilocybin and Derivatives in Tryptophan Synthase-Enhanced Reactions". Chemistry: A European Journal 24 (40): 10028–10031. May 2018. doi:10.1002/chem.201801047. PMID 29750381.
- ↑ 6.0 6.1 "Psychedelic effects of psilocybin correlate with serotonin 2A receptor occupancy and plasma psilocin levels". Neuropsychopharmacology 44 (7): 1328–1334. June 2019. doi:10.1038/s41386-019-0324-9. PMID 30685771.
- ↑ 7.0 7.1 7.2 "Recreational use, analysis and toxicity of tryptamines". Current Neuropharmacology 13 (1): 26–46. January 2015. doi:10.2174/1570159X13666141210222409. PMID 26074742.
- ↑ "L-tryptophan decarboxylase". SWISS-MODEL Repository. Expasy, Swiss Institute of Bioinformatics (SIB). https://swissmodel.expasy.org/repository/uniprot/P0DPA6?template=6l06.1.A&range=172-401.
- ↑ 9.0 9.1 9.2 9.3 "Horizontal gene cluster transfer increased hallucinogenic mushroom diversity". Evolution Letters 2 (2): 88–101. April 2018. doi:10.1002/evl3.42. PMID 30283667.
- ↑ 10.0 10.1 "Convergent evolution of psilocybin biosynthesis by psychedelic mushrooms" (in en). bioRxiv: 374199. 2018-07-27. doi:10.1101/374199. https://www.biorxiv.org/content/10.1101/374199v2.
- ↑ "A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation". Science 318 (5858): 1913–1916. December 2007. doi:10.1126/science.1146954. PMID 18096805. Bibcode: 2007Sci...318.1913H.
- ↑ "The Trace Fossil Record of Eusociality in Ants and Termites" (in en). Ichnoentomology: Insect Traces in Soils and Paleosols. Topics in Geobiology. 37. Cham: Springer International Publishing. 2017. pp. 285–312. doi:10.1007/978-3-319-28210-7_12. ISBN 978-3-319-28210-7.
 |

