Biology:Human Y-chromosome DNA haplogroup
| Part of a series on |
| Genetic genealogy |
|---|
| Concepts |
| Related topics |
In human genetics, a human Y-chromosome DNA haplogroup is a haplogroup defined by mutations in the non-recombining portions of DNA from the male-specific Y chromosome (called Y-DNA). Many people within a haplogroup share similar numbers of short tandem repeats (STRs) and types of mutations called single-nucleotide polymorphisms (SNPs).[2]
The human Y-chromosome accumulates roughly two mutations per generation.[3] Y-DNA haplogroups represent major branches of the Y-chromosome phylogenetic tree that share hundreds or even thousands of mutations unique to each haplogroup.
The Y-chromosomal most recent common ancestor (Y-MRCA, informally known as Y-chromosomal Adam) is the most recent common ancestor (MRCA) from whom all currently living humans are descended patrilineally. Y-chromosomal Adam is estimated to have lived roughly 236,000 years ago in Africa[citation needed]. By examining other bottlenecks most Eurasian men (men from populations outside of Africa) are descended from a man who lived in Africa 69,000 years ago (Haplogroup CT). Other major bottlenecks occurred about 50,000 and 5,000 years ago and subsequently the ancestry of most Eurasian men can be traced back to four ancestors who lived 50,000 years ago, who were descendants of African (E-M168).[4][5][6][clarification needed]
Naming convention
Y-DNA haplogroups are defined by the presence of a series of Y-DNA SNP markers. Subclades are defined by a terminal SNP, the SNP furthest down in the Y-chromosome phylogenetic tree.[7][8] The Y Chromosome Consortium (YCC) developed a system of naming major Y-DNA haplogroups with the capital letters A through T, with further subclades named using numbers and lower case letters (YCC longhand nomenclature). YCC shorthand nomenclature names Y-DNA haplogroups and their subclades with the first letter of the major Y-DNA haplogroup followed by a dash and the name of the defining terminal SNP.[9]
Y-DNA haplogroup nomenclature is changing over time to accommodate the increasing number of SNPs being discovered and tested, and the resulting expansion of the Y-chromosome phylogenetic tree. This change in nomenclature has resulted in inconsistent nomenclature being used in different sources.[2] This inconsistency, and increasingly cumbersome longhand nomenclature, has prompted a move toward using the simpler shorthand nomenclature.
Phylogenetic structure
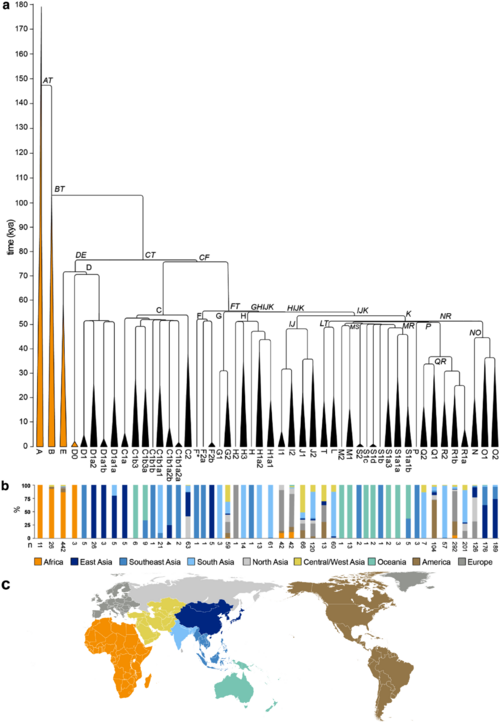
- Phylogenetic tree of Y-DNA haplogroups [10]
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Major Y-DNA haplogroups
Haplogroups A and B
Haplogroup A is the NRY (non-recombining Y) macrohaplogroup from which all modern paternal haplogroups descend. It is sparsely distributed in Africa, being concentrated among Khoisan populations in the southwest and Nilotic populations toward the northeast in the Nile Valley. BT is a subclade of haplogroup A, more precisely of the A1b clade (A2-T in Cruciani et al. 2011), as follows:
- Haplogroup A
- Haplogroup A00
- Haplogroup A0 (formerly also A1b)
- Haplogroup A1 (also A1a-T)
- Haplogroup A1a (M31)
- Haplogroup A1b (also A2-T; P108, V221)
- Haplogroup A1b1a1 (also A2; M14)
- Haplogroup A1b1b (also A3; M32)
- Haplogroup BT (M91, M42, M94, M139, M299)
- Haplogroup B (M60)
- Haplogroup CT
Haplogroup CT (P143)
The defining mutations separating CT (all haplogroups except for A and B) are M168 and M294. The site of origin is likely in Africa. Its age has been estimated at approximately 88,000 years old,[11][12] and more recently at around 100,000[13] or 101,000 years old.[14]
Haplogroup C (M130)
- Haplogroup C (M130, M216) Found in Asia, Oceania, and North America
- Haplogroup C1 (F3393/Z1426)
- Haplogroup C1a (CTS11043)
- Haplogroup C1a1 (M8, M105, M131) Found with low frequency in Japan
- Haplogroup C1a2 (V20) Found with low frequency in Europe, Armenians, Algeria, and Nepal
- Haplogroup C1b (F1370, Z16480)
- Haplogroup C1b1 (AM00694/K281)
- Haplogroup C1b1a (B66/Z16458)
- Haplogroup C1b1a1 (M356) Found with low frequency in South Asia, Southwest Asia, and northern China
- Haplogroup C1b1a2 (B65)
- Haplogroup C1b1a2a (B67) Found among Lebbo' people in Borneo, Indonesia
- Haplogroup C1b1a2b (F725) Found among Han Chinese (Guangdong, Hunan, and Shaanxi), Dai people (Yunnan), Murut people (Brunei), Malay people (Singapore), and Aeta people (Philippines)
- Haplogroup C1b1a3 (Z16582) Found with low frequency in Saudi Arabia and Iraq
- Haplogroup C1b1b (B68) Found among Dusun people (Brunei)
- Haplogroup C1b1a (B66/Z16458)
- Haplogroup C1b2 (C-Z16582)
- Haplogroup C1b3 (B477/Z31885)
- Haplogroup C1b3a (M38) Found in Indonesia, New Guinea, Melanesia, Micronesia, and Polynesia
- Haplogroup C1b3b (M347, P309) Found among the indigenous peoples in Australia
- Haplogroup C1b1 (AM00694/K281)
- Haplogroup C1a (CTS11043)
- Haplogroup C2 (M217, P44) Found throughout Eurasia and North America, but especially among Mongols, Kazakhs, Tungusic peoples, Paleosiberians, and Na-Dené-speaking peoples
- Haplogroup C1 (F3393/Z1426)
Haplogroup D (CTS3946)
- Haplogroup D (CTS3946)
- Haplogroup D1 (M174) Found in Japan , China (especially Tibet), the Andaman Islands
- Haplogroup D1a (CTS11577)
- Haplogroup D1a1 (Z27276, Z27283, Z29263)
- Haplogroup D1a1a (M15) Found mainly in Tibetans, Qiangic peoples, Yi, and Hmong-Mien peoples
- Haplogroup D1a1b (P99) Found mainly in Tibetans, Qiangic peoples, Naxi, and Turkic peoples
- Haplogroup D1a2 (M55, M57, M64.1, M179, P12, P37.1, P41.1 (M359.1), 12f2.2) Found mainly in Japan
- Haplogroup D1a3 (Y34637) Found in Andamanese peoples (Onge, Jarawa)
- Haplogroup D1a1 (Z27276, Z27283, Z29263)
- Haplogroup D1b (L1366, L1378, M226.2) Found in Mactan Island, Philippines
- Haplogroup D1a (CTS11577)
- Haplogroup D2 (A5580.2) Found in Nigeria, Saudi Arabia and Syria
Haplogroup E (M96)
- Haplogroup E (M40, M96) Found in Africa and parts of the Middle East and Europe
- Haplogroup E1 (P147)
- Haplogroup E1a (M33, M132) formerly E1
- Haplogroup E1b (P177)
- Haplogroup E1b1 (P2, DYS391p); formerly E3
- Haplogroup E1b1a (V38)
- Haplogroup E1b1a1 (M2) Found in Africa, especially among Niger–Congo-speaking populations.; formerly E3a
- Haplogroup E1b1a2 (M329) Found in Africa, especially in Ethiopia among Omotic-speaking populations.; formerly E3*
- Haplogroup E1b1b (M215)
- Haplogroup E1b1b1 (M35) Found in Horn of Africa, North Africa, the Middle East, and Europe (especially in areas near the Mediterranean and the Balkans); formerly E3b
- Haplogroup E1b1a (V38)
- Haplogroup E1b1 (P2, DYS391p); formerly E3
- Haplogroup E2 (M75)
- Haplogroup E1 (P147)
Haplogroup F (M89)
The groups descending from haplogroup F are found in some 90% of the world's population, but almost exclusively outside of sub-Saharan Africa.
F xG,H,I,J,K is rare in modern populations and peaks in South Asia, especially Sri Lanka.[10] It also appears to have long been present in South East Asia; it has been reported at rates of 4–5% in Sulawesi and Lembata. One study, which did not comprehensively screen for other subclades of F-M89 (including some subclades of GHIJK), found that Indonesian men with the SNP P14/PF2704 (which is equivalent to M89), comprise 1.8% of men in West Timor, 1.5% of Flores 5.4% of Lembata 2.3% of Sulawesi and 0.2% in Sumatra.[15][16] F* (F xF1,F2,F3) has been reported among 10% of males in Sri Lanka and South India, 5% in Pakistan, as well as lower levels among the Tamang people (Nepal), and in Iran. F1 (P91), F2 (M427) and F3 (M481; previously F5) are all highly rare and virtually exclusive to regions/ethnic minorities in Sri Lanka, India, Nepal, South China, Thailand, Burma, and Vietnam. In such cases, however, the possibility of misidentification is considered to be relatively high and some may belong to misidentified subclades of Haplogroup GHIJK.[17]
Haplogroup G (M201)
Haplogroup G (M201) originated some 48,000 years ago and its most recent common ancestor likely lived 26,000 years ago in the Middle East. It spread to Europe with the Neolithic Revolution.
It is found in many ethnic groups in Eurasia; most common in the Caucasus, Iran, Anatolia and the Levant. Found in almost all European countries, but most common in Gagauzia, southeastern Romania, Greece, Italy, Spain , Portugal, Tyrol, and Bohemia with highest concentrations on some Mediterranean islands; uncommon in Northern Europe.[18][19]
G-M201 is also found in small numbers in northwestern China and India , Bangladesh, Pakistan , Sri Lanka, Malaysia, and North Africa.
- Haplogroup G1
- Haplogroup G2
- Haplogroup G2a
- Haplogroup G2a1
- Haplogroup G2a2
- Haplogroup G2a3
- Haplogroup G2a3a
- Haplogroup G2a3b
- Haplogroup G2a3b1
- Haplogroup G2b
- Haplogroup G2c (formerly Haplogroup G5)
- Haplogroup G2c1
- Haplogroup G2a
Haplogroup H (M69)
Haplogroup H (M69) probably emerged in Southern Central Asia, South Asia or West Asia, about 48,000 years BP, and remains largely prevalent there in the forms of H1 (M69) and H3 (Z5857). Its sub-clades are also found in lower frequencies in Iran, Central Asia, across the middle-east, and the Arabian peninsula.
However, H2 (P96) is present in Europe since the Neolithic and H1a1 (M82) spread westward in the Medieval era with the migration of the Roma people.
Haplogroup I (M170)
Haplogroup I (M170, M258) is found mainly in Europe and the Caucasus.
- Haplogroup I1 Nordid/Nordic Europids (M253) Found mainly in northern Europe
- Haplogroup I2 Dinarid/Dinaric Europids (P215) Found mainly in Balkans, southeast Europe and Sardinia save for I2B1 (m223) which is found at a moderate frequency in Western, Central, and Northern Europe.
Haplogroup J (M304)
Haplogroup J (M304, S6, S34, S35) is found mainly in the Middle East, Caucasus and South-East Europe.
- Haplogroup J* (J-M304*) is rare outside the island of Socotra.
- Haplogroup J1 Semitid/Bedouinid Arabids (M267) are associated with Northeast Caucasian peoples in Dagestan and Semitic languages speaking people in the Middle East, Ethiopia, and North Africa and also found in Mediterranean Europe in smaller frequencies much like haplogroup T.
- Haplogroup J2 Syrid/Nahrainid Arabids (M172) is found mainly in the Semitic-speaking peoples, Anatolia, Greece, the Balkans, Italy, Iran, the Caucasus, South Asia, and Central Asia.
Haplogroup K (M9)
Haplogroup K (M9) is spread all over Eurasia, Oceania and among Native Americans.
K(xLT,K2a,K2b) – that is, K*, K2c, K2d or K2e – is found mainly in Melanesia, Aboriginal Australians, India , Polynesia and Island South East Asia.
Haplogroups L and T (K1)
Haplogroup L (M20) is found in South Asia, Central Asia, South-West Asia, and the Mediterranean.
Haplogroup T (M184, M70, M193, M272) is found at high levels in the Horn of Africa (mainly Cushitic-speaking peoples), parts of South Asia, the Middle East, and the Mediterranean. T-M184 is also found in significant minorities of Sciaccensi, Stilfser, Egyptians, Omanis, Sephardi Jews,[20] Ibizans (Eivissencs), and Toubou. It is also found at low frequencies in other parts of the Mediterranean and South Asia.
Haplogroup K2 (K-M526)
The only living males reported to carry the basal paragroup K2* are indigenous Australians. Major studies published in 2014 and 2015 suggest that up to 27% of Aboriginal Australian males carry K2*, while others carry a subclade of K2.
Haplogroups K2a, K2a1, NO & NO1
Haplogroup N
Haplogroup N (M231) is found in northern Eurasia, especially among speakers of the Uralic languages.
Haplogroup N possibly originated in eastern Asia and spread both northward and westward into Siberia, being the most common group found in some Uralic-speaking peoples.
Haplogroup O
Haplogroup O (M175) is found with its highest frequency in East Asia and Southeast Asia, with lower frequencies in the South Pacific, Central Asia, South Asia, and islands in the Indian Ocean (e.g. Madagascar, the Comoros).
- Haplogroup O1 (F265/M1354, CTS2866, F75/M1297, F429/M1415, F465/M1422)
- Haplogroup O1a (M119, CTS31, F589/Page20, L246, L466) Found in eastern, central, and southern Mainland China, Taiwan, and Southeast Asia, especially among Austronesian and Tai–Kadai peoples
- Haplogroup O1b (P31, M268)
- Haplogroup O1b1 (M95) Found in Japan , China, Taiwan, Southeast Asia, and the Indian subcontinent, especially among Austroasiatic- and Tai–Kadai-speaking peoples, Malays, and Indonesians
- Haplogroup O1b2 (SRY465, M176) Found in Japan, Korea, Manchuria, and Southeast Asia
- Haplogroup O2 (M122) Found throughout East Asia, Southeast Asia, and Austronesia including Polynesia
Haplogroups K2b1, M & S
No examples of the basal paragroup K2b1* have been identified. Males carrying subclades of K2b1 are found primarily among Papuan peoples, Micronesian peoples, indigenous Australians, and Polynesians.
Its primary subclades are two major haplogroups:
- Haplogroup S (B254) also known as K2b1a: found in the highlands of Papua New Guinea and;
- Haplogroup M (P256) also known as K2b1b: found in New Guinea and Melanesia.
Haplogroup P (K2b2)
Haplogroup P (P295) has two primary branches: P1 (P-M45) and the extremely rare P2 (P-B253).[21]
P*, P1* and P2 are found together only on the island of Luzon in the Philippines .[21] In particular, P* and P1* are found at significant rates among members of the Aeta (or Agta) people of Luzon.[22] While, P1* is now more common among living individuals in Eastern Siberia and Central Asia, it is also found at low levels in mainland South East Asia and South Asia. Considered together, these distributions tend to suggest that P* emerged from K2b in South East Asia.[22][23]
P1 is also the parent node of two primary clades:
- Haplogroup Q (Q-M242) and;
- Haplogroup R (R-M207). These share the common marker M45 in addition to at least 18 other SNPs.
Haplogroup Q (MEH2, M242, P36) found in Siberia and the Americas Haplogroup R (M207, M306): found in Europe, West Asia, Central Asia, and South Asia
Haplogroup Q M242
Q is defined by the SNP M242. It is believed to have arisen in Central Asia approximately 32,000 years ago.[24][25] The subclades of Haplogroup Q with their defining mutation(s), according to the 2008 ISOGG tree[26] are provided below. ss4 bp, rs41352448, is not represented in the ISOGG 2008 tree because it is a value for an STR. This low frequency value has been found as a novel Q lineage (Q5) in Indian populations[27]
The 2008 ISOGG tree
- Q (M242)
- Q*
- Q1 (P36.2)
- Q1*
- Q1a (MEH2)
- Q1a*
- Q1a1 (M120, M265/N14) Found with low frequency among Bhutanese,[28] Dungans, Han Chinese, Japanese, Koreans, Mongolians,[29] Naxi, and Tibetans[30][31]
- Q1a2 (M25, M143) Found at low to moderate frequency among some populations of Southwest Asia, Central Asia, and Siberia
- Q1a3 (M346)
- Q1a3* Found at low frequency in Pakistan , India , and Tibet
- Q1a3a (M3) Typical of indigenous peoples of the Americas
- Q1a3a*
- Q1a3a1 (M19) Found among some indigenous peoples of South America, such as the Ticuna and the Wayuu[32]
- Q1a3a2 (M194)
- Q1a3a3 (M199, P106, P292)
- Q1a4 (P48)
- Q1a5 (P89)
- Q1a6 (M323) Found in a significant minority of Yemeni Jews
- Q1b (M378) Found at low frequency among samples of Hazara and Sindhis
Haplogroup R (M207)
Haplogroup R is defined by the SNP M207. The bulk of Haplogroup R is represented in the descendant subclade R1 (M173), which originated on the Siberia. R1 has two descendant subclades: R1a and R1b.
R1a is associated with the proto-Indo-Iranian and Balto-Slavic peoples, and is now found primarily in Central Asia, South Asia, and Eastern Europe.
Haplogroup R1b is the dominant haplogroup of Western Europe and is also found sparsely distributed among various peoples of Asia and Africa. Its subclade R1b1a2 (M269) is the haplogroup that is most commonly found among modern Western European populations, and has been associated with the Italo-Celtic and Germanic peoples.
- Haplogroup R1 (M173) Found throughout western Eurasia
- Haplogroup R1a (M420) Found in Central Asia, South Asia, and Central, Northern and Eastern Europe, Balkans
- Haplogroup R1b (M343) Found in Western Europe, West Asia, Central Asia, North Africa, and northern Cameroon
- Haplogroup R2 (M124) Found in South Asia, Caucasus, Central Asia, and Eastern Europe
Chronological development of haplogroups
| Haplogroup | Possible time of origin | Possible place of origin | Possible TMRCA[33][12] |
|---|---|---|---|
| A00 | 235,900[5] or 275,000 years ago[34] | Africa[35] | 235,900 years ago |
| BT | 130,700 years ago[5] | Africa | 88,000 years ago |
| CT | 88,000[5] or 101-100,000 years ago[13][14] | Africa | 68,500 years ago |
| E | 65,200,[5] 69,000,[36] or 73,000 years ago[37] | East Africa[38] or Asia[15] | 53,100 years ago |
| F | 65,900 years ago[5] | Eurasia | 48,800 years ago |
| G | 48,500 years ago[5] | Middle East | 26,200 years ago |
| IJ | 47,200 years ago[5] | Middle East | 42,900 years ago |
| K | 47,200 years ago[5] | Asia | 45,400 years ago |
| P | 45,400 years ago[5] | Asia | 31,900 years ago |
| J | 42,900 years ago[5][39] | Middle East | 31,600 years ago |
| I | 42,900 years ago[5] | Europe | 27,500 years ago |
| E-M215 (E1b1b) | 42,300 years ago[5][40] | East Africa | 34,800 years ago |
| E-V38 (E1b1a) | 42,300 years ago[5][40] | East Africa | 40,100 years ago |
| N | 36,800 years ago[5][41] | Asia | 22,100 years ago |
| E1b1b-M35 | 34,800 years ago[5][40] | East Africa | 24,100 years ago |
| R | 31,900 years ago[5] | Asia | 28,200 years ago |
| J-M267 (J1) | 31,600 years ago[5][39] | Middle East | 18,500 years ago |
| J-M172 (J2) | 31,600 years ago[5][39] | Middle East | 27,800 years ago[5][42] |
| R-M173 (R1) | 28,200 years ago[5] | Asia | 22,800 years ago |
| I-M253 (I1) | 27,500 years ago[5][43][44] | Europe | 4,600 years ago |
| I-M438 (I2) | 27,500 years ago[5][44] | Europe | 21,800 years ago |
| R-M420 (R1a) | 22,800 years ago[5][45] | Eurasia | 18,300 years ago |
| R-M343 (R1b) | 22,800 years ago[5][46] | Eurasia[47] | 20,400 years ago |
| I2-L460 (I2a) | 21,800 years ago[5][48] | Europe | 21,100 years ago |
| I2a-P37 | 21,100 years ago[5][43][49] | Europe | 18,500 years ago |
| E1b1b-M78 | 19,800 years ago[5][40][50] | Northeast Africa[50] | 13,400 years ago[5][50] |
| I2a-M423 | 18,500 years ago[5][49] | Europe | 13,500 years ago |
| I2a-M223 | 17,400 years ago[5] | Europe | 12,100 years ago |
| N1c-M178 | 14,200 years ago[5][41] | Asia | 11,900 years ago |
| R1a-M17 | 14,100 years ago[5][45][51] | Eastern Europe | 8,500 years ago |
| R1b-M269 | 13,300 years ago[5] | Eastern Europe | 6,400 years ago[52] |
| E1b1b-V12 | 11,800 years ago[5][50] | North Africa | 9,900 years ago |
| E-U175 (E1b1a8) | 9,200 years ago[5][40] | East Africa | 8,500 years ago |
| E1b1b-V13 | 8,100 years ago[5][50] | Southern Europe | 4,800 years ago |
| E-M191 (E1b1a7) | 7,400 years ago[5][40] | East Africa | 6,400 years ago |
| E-U174 (E1b1a-U174) | 6,400 years ago[5][40] | East Africa | 5,300 years ago |
| R1b-L151 | 5,800 years ago[5] | Eastern Europe | 4,800 years ago |
| R1a-Z280 | 5,000 years ago[5] | Eastern Europe | 4,600 years ago[53] |
| R1a-M458 | 4,700 years ago[5] | Eastern Europe | 4,700 years ago[53] |
See also
- List of Y-chromosome haplogroups in populations of the world
- Y-DNA haplogroups in populations of Europe
- Genetic history of Europe
- List of Y-DNA single-nucleotide polymorphisms
- List of Y-STR markers
- Human mitochondrial DNA haplogroup
- * (haplogroup)
- Molecular phylogeny
- Genetic genealogy
- Genealogical DNA test
- Conversion table for Y chromosome haplogroups
References
- ↑ Hallast P, Agdzhoyan A, Balanovsky O, Xue Y, Tyler-Smith C (2020). "A Southeast Asian origin for present-day non-African human Y chromosomes.". Hum Genet 140 (2): 299–307. doi:10.1007/s00439-020-02204-9. PMID 32666166.
- ↑ 2.0 2.1 "Understanding Haplogroups: How are the haplogroups named?". Family Tree DNA. http://www.familytreedna.com/understanding-haplogroups.aspx.
- ↑ Dolgin, Elie (2009). "Human mutation rate revealed". Nature. doi:10.1038/news.2009.864. http://www.nature.com/news/2009/090827/full/news.2009.864.html. Retrieved 18 September 2017. "one mutation in every 30 million base pairs"
- ↑ Karmin (2015). "A recent bottleneck of Y chromosome diversity coincides with a global change in culture". Genome Research 25 (4): 459–66. doi:10.1101/gr.186684.114. PMID 25770088. "we date the Y-chromosomal most recent common ancestor (MRCA) in Africa at 254 (95% CI 192–307) kya and detect a cluster of major non-African founder haplogroups in a narrow time interval at 47–52 kya, consistent with a rapid initial colonization model of Eurasia and Oceania after the out-of-Africa bottleneck. In contrast to demographic reconstructions based on mtDNA, we infer a second strong bottleneck in Y-chromosome lineages dating to the last 10 ky. We hypothesize that this bottleneck is caused by cultural changes affecting variance of reproductive success among males."
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 5.17 5.18 5.19 5.20 5.21 5.22 5.23 5.24 5.25 5.26 5.27 5.28 5.29 5.30 5.31 5.32 5.33 5.34 5.35 5.36 5.37 5.38 5.39 5.40 5.41 "YFull YTree". YFull. https://www.yfull.com/tree.
- ↑ "Something Weird Happened to Men 7,000 Years Ago, And We Finally Know Why". 31 May 2018. https://www.sciencealert.com/neolithic-y-chromosome-bottleneck-warring-patrilineal-clans. "Around 7000 years ago - all the way back in the Neolithic - something really peculiar happened to human genetic diversity. Over the next 2,000 years, and seen across Africa, Europe and Asia, the genetic diversity of the Y chromosome collapsed, becoming as though there was only one man for every 17 women."
- ↑ "Understanding Results: Y-DNA Single Nucleotide Polymorphism (SNP): What is a Y-chromosome DNA (Y-DNA) haplogroup?". Family Tree DNA. http://www.familytreedna.com/faq/answers.aspx?id=26#314. "Y-chromosome DNA (Y-DNA) haplogroups are the major branches on the human paternal family tree. Each haplogroup has many subbranches. These are subclades."
- ↑ "myFTDNA 2.0 User Guide: Y-DNA: What is the Y-DNA – Matches page?". Family Tree DNA. http://www.familytreedna.com/faq/answers.aspx?id=48#1990. "A terminal SNP determines the terminal (final) subbranch on the Y-DNA Tree to which someone belongs."
- ↑ "Understanding Results: Y-DNA Single Nucleotide Polymorphism (SNP): How are haplogroups and their subclades named?". Family Tree DNA. http://www.familytreedna.com/faq/answers.aspx?id=26#318.
- ↑ 10.0 10.1 Copyright 2015 ISOGG. "ISOGG 2015 Y-DNA Haplogroup Tree Trunk". isogg.org. http://www.isogg.org/tree/ISOGG_YDNATreeTrunk.html.
- ↑ Underhill and Kivisild; Kivisild, T (2007). "Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations". Annu. Rev. Genet. 41 (1): 539–64. doi:10.1146/annurev.genet.41.110306.130407. PMID 18076332.
- ↑ 12.0 12.1 Karafet, TM; Mendez, FL; Meilerman, MB; Underhill, PA; Zegura, SL; Hammer, MF (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–38. doi:10.1101/gr.7172008. PMID 18385274.
- ↑ 13.0 13.1 "A recent bottleneck of Y chromosome diversity coincides with a global change in culture". Genome Research 25 (4): 459–66. April 2015. doi:10.1101/gr.186684.114. PMID 25770088.
- ↑ 14.0 14.1 "A Rare Deep-Rooting D0 African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa". Genetics 212 (4): 1421–28. June 2019. doi:10.1534/genetics.119.302368. PMID 31196864.
- ↑ 15.0 15.1 Chiaroni, Jacques; Underhill, Peter A.; Cavalli-Sforza, Luca L. (1 December 2009). "Y chromosome diversity, human expansion, drift, and cultural evolution". Proceedings of the National Academy of Sciences of the United States of America 106 (48): 20174–79. doi:10.1073/pnas.0910803106. PMID 19920170. Bibcode: 2009PNAS..10620174C.
- ↑ Tumonggor, Meryanne K (2014). "Isolation, contact and social behavior shaped genetic diversity in West Timor". Journal of Human Genetics 59 (9): 494–503. doi:10.1038/jhg.2014.62. PMID 25078354.
- ↑ This was, for instance, the case with the original subclade F3 (M96), which has since been renamed Haplogroup H2.
- ↑ Passarino, Giuseppe; Cavalleri, Gianpiero L; Lin, Alice A; Cavalli-Sforza, LL; Børresen-Dale, AL; Underhill, PA (2002). "Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms". European Journal of Human Genetics 10 (9): 521–29. doi:10.1038/sj.ejhg.5200834. PMID 12173029.
- ↑ Karlsson, Andreas O; Wallerström, Thomas; Götherström, Anders; Holmlund, Gunilla (2006). "Y-chromosome diversity in Sweden – A long-time perspective". European Journal of Human Genetics 14 (8): 963–70. doi:10.1038/sj.ejhg.5201651. PMID 16724001.
- ↑ Nogueiro, Inês (2009). "Phylogeographic analysis of paternal lineages in NE Portuguese Jewish communities". American Journal of Physical Anthropology 141 (3): 373–81. doi:10.1002/ajpa.21154. PMID 19918998.
- ↑ 21.0 21.1 ISOGG, 2016, Y-DNA Haplogroup P and its Subclades – 2016 (20 June 2016).
- ↑ 22.0 22.1 Tumonggor, Meryanne K; Karafet, Tatiana M; Downey, Sean; Lansing, J Stephen; Norquest, Peter; Sudoyo, Herawati; Hammer, Michael F; Cox, Murray P (31 July 2014). "Isolation, contact and social behavior shaped genetic diversity in West Timor". Journal of Human Genetics 59 (9): 494–503. doi:10.1038/jhg.2014.62. PMID 25078354.
- ↑ Tatiana M Karafet (2015). "Improved phylogenetic resolution and rapid diversification of Y-chromosome haplogroup K-M526 in Southeast Asia". European Journal of Human Genetics 23 (3): 369–73. doi:10.1038/ejhg.2014.106. PMID 24896152.
- ↑ Fagundes, Nelson J.R.; Ricardo Kanitz; Roberta Eckert; Ana C.S. Valls; Mauricio R. Bogo; Francisco M. Salzano; David Glenn Smith; Wilson A. Silva et al. (2008). "Mitochondrial Population Genomics Supports a Single Pre-Clovis Origin with a Coastal Route for the Peopling of the Americas". American Journal of Human Genetics 82 (3): 583–92. doi:10.1016/j.ajhg.2007.11.013. PMID 18313026. PMC 2427228. http://www.familytreedna.com/pdf/Fagundes-et-al.pdf. Retrieved 2013-05-22. "Since the first studies, it has been found that extant Native American populations exhibit almost exclusively five "mtDNA haplogroups" (A–D and X)6 classified in the autochthonous haplogroups A2, B2, C1, D1, and X2a.7 Haplogroups A–D are found all over the New World and are frequent in Asia, supporting a northeastern Asian origin of these lineages".
- ↑ Zegura, S. L.; Karafet, TM; Zhivotovsky, LA; Hammer, MF (2003). "High-Resolution SNPs and Microsatellite Haplotypes Point to a Single, Recent Entry of Native American Y Chromosomes into the Americas". Molecular Biology and Evolution 21 (1): 164–75. doi:10.1093/molbev/msh009. PMID 14595095.
- ↑ "Y-DNA Haplogroup Tree 2010". International Society of Genetic Genealogy. http://www.isogg.org/tree/.
- ↑ Sharma, Swarkar; Rai, Ekta; Bhat, Audesh K; Bhanwer, Amarjit S; Bamezaicorresponding, Rameshwar NK (2007). "A novel subgroup Q5 of human Y-chromosomal haplogroup Q in India". BMC Evol Biol 7: 232. doi:10.1186/1471-2148-7-232. PMID 18021436.
- ↑ Hallast, P.; Batini, C.; Zadik, D.; Maisano Delser, P.; Wetton, J. H.; Arroyo-Pardo, E.; Cavalleri, G. L.; De Knijff, P. et al. (2015). "The Y-Chromosome Tree Bursts into Leaf: 13,000 High-Confidence SNPs Covering the Majority of Known Clades". Molecular Biology and Evolution 32 (3): 661–73. doi:10.1093/molbev/msu327. PMID 25468874.
- ↑ Di Cristofaro, Julie; Pennarun, Erwan; Mazières, Stéphane; Myres, Natalie M.; Lin, Alice A.; Temori, Shah Aga; Metspalu, Mait; Metspalu, Ene et al. (2013). "Afghan Hindu Kush: Where Eurasian Sub-Continent Gene Flows Converge". PLOS ONE 8 (10): e76748. doi:10.1371/journal.pone.0076748. PMID 24204668. Bibcode: 2013PLoSO...876748D.
- ↑ "Genetic evidence supports demic diffusion of Han culture". Nature 431 (7006): 302–05. September 2004. doi:10.1038/nature02878. PMID 15372031. Bibcode: 2004Natur.431..302W.
- ↑ "The Eurasian heartland: a continental perspective on Y-chromosome diversity". Proc. Natl. Acad. Sci. U.S.A. 98 (18): 10244–49. August 2001. doi:10.1073/pnas.171305098. PMID 11526236. Bibcode: 2001PNAS...9810244W.
- ↑ "Y-chromosome evidence for differing ancient demographic histories in the Americas". Am. J. Hum. Genet. 73 (3): 524–39. September 2003. doi:10.1086/377588. PMID 12900798.
- ↑ Batini, Chiara; Hallast, Pille; Zadik, Daniel; Delser, Pierpaolo Maisano; Benazzo, Andrea; Ghirotto, Silvia; Arroyo-Pardo, Eduardo; Cavalleri, Gianpiero L. et al. (2015). "Large-scale recent expansion of European patrilineages shown by population resequencing". Nature Communications 6: 7152. doi:10.1038/ncomms8152. PMID 25988751. Bibcode: 2015NatCo...6.7152B.
- ↑ Mendez, L. (2016). "The Divergence of Neandertal and Modern Human Y Chromosomes". The American Journal of Human Genetics 98 (4): 728–34. doi:10.1016/j.ajhg.2016.02.023. PMID 27058445.
- ↑ "The father of all men is 340,000 years old". https://www.newscientist.com/article/dn23240-the-father-of-all-men-is-340000-years-old/.
- ↑ "A recent bottleneck of Y chromosome diversity coincides with a global change in culture". Genome Research 25 (4): 459–466. April 2015. doi:10.1101/gr.186684.114. PMID 25770088.
- ↑ "A Rare Deep-Rooting D0 African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa". Genetics 212 (4): 1421–1428. June 2019. doi:10.1534/genetics.119.302368. PMID 31196864.
- ↑ Semino, Ornella; Magri, Chiara; Benuzzi, Giorgia; Lin, Alice A.; Al-Zahery, Nadia; Battaglia, Vincenza; MacCioni, Liliana; Triantaphyllidis, Costas et al. (2004). "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area". The American Journal of Human Genetics 74 (5): 1023–34. doi:10.1086/386295. PMID 15069642.
- ↑ 39.0 39.1 39.2 Semino, OExpression error: Unrecognized word "etal". (May 2004). "Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area". Am. J. Hum. Genet. 74 (5): 1023–34. doi:10.1086/386295. PMID 15069642.
- ↑ 40.0 40.1 40.2 40.3 40.4 40.5 40.6 Trombetta, Beniamino; d'Atanasio, Eugenia; Massaia, Andrea; Ippoliti, Marco; Coppa, Alfredo; Candilio, Francesca; Coia, Valentina; Russo, Gianluca et al. (2015). "Phylogeographic Refinement and Large Scale Genotyping of Human y Chromosome Haplogroup e Provide New Insights into the Dispersal of Early Pastoralists in the African Continent". Genome Biology and Evolution 7 (7): 1940–50. doi:10.1093/gbe/evv118. PMID 26108492.
- ↑ 41.0 41.1 Shi, Hong; Qi, Xuebin; Zhong, Hua; Peng, Yi; Zhang, Xiaoming; Ma, Runlin Z.; Su, Bing (2013). "Genetic Evidence of an East Asian Origin and Paleolithic Northward Migration of Y-chromosome Haplogroup N". PLOS ONE 8 (6): e66102. doi:10.1371/journal.pone.0066102. PMID 23840409. Bibcode: 2013PLoSO...866102S.
- ↑ Batini, Chiara; Hallast, Pille; Zadik, Daniel; Delser, Pierpaolo Maisano; Benazzo, Andrea; Ghirotto, Silvia; Arroyo-Pardo, Eduardo; Cavalleri, Gianpiero L. et al. (2015). "Large-scale recent expansion of European patrilineages shown by population resequencing". Nature Communications 6: 7152. doi:10.1038/ncomms8152. PMID 25988751. Bibcode: 2015NatCo...6.7152B.
- ↑ 43.0 43.1 Rootsi, Siiri (2004). "Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe". American Journal of Human Genetics 75 (1): 128–37. doi:10.1086/422196. PMID 15162323. PMC 1181996. http://www.familytreedna.com/pdf/DNA.RootsiHaplogroupISpread.pdf. Retrieved 2016-05-04.
- ↑ 44.0 44.1 P.A. Underhill, N.M. Myres, S. Rootsi, C.T. Chow, A.A. Lin, R.P. Otillar, R. King, L.A. Zhivotovsky, O. Balanovsky, A. Pshenichnov, K.H. Ritchie, L.L. Cavalli-Sforza, T. Kivisild, R. Villems, S.R. Woodward, New Phylogenetic Relationships for Y-chromosome Haplogroup I: Reappraising its Phylogeography and Prehistory, in P. Mellars, K. Boyle, O. Bar-Yosef and C. Stringer (eds.), Rethinking the Human Evolution (2007), pp. 33–42.
- ↑ 45.0 45.1 Sharma, Swarkar; Rai, Ekta; Sharma, Prithviraj; Jena, Mamata; Singh, Shweta; Darvishi, Katayoon; Bhat, Audesh K.; Bhanwer, A J S. et al. (2009). "The Indian origin of paternal haplogroup R1a1* substantiates the autochthonous origin of Brahmins and the caste system". Journal of Human Genetics 54 (1): 47–55. doi:10.1038/jhg.2008.2. PMID 19158816.
- ↑ ftDNA
- ↑ Myres2010
- ↑ Jones, Eppie R.; Gonzalez-Fortes, Gloria; Connell, Sarah; Siska, Veronika; Eriksson, Anders; Martiniano, Rui; McLaughlin, Russell L.; Gallego Llorente, Marcos et al. (2015). "Upper Palaeolithic genomes reveal deep roots of modern Eurasians". Nature Communications 6: 8912. doi:10.1038/ncomms9912. PMID 26567969. Bibcode: 2015NatCo...6.8912J.
- ↑ 49.0 49.1 "Mesolithic Western Eurasian DNA". Ancestral Journeys. http://www.ancestraljourneys.org/mesolithicdna.shtml.
- ↑ 50.0 50.1 50.2 50.3 50.4 Cruciani (2007), "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12", Molecular Biology and Evolution 24 (6): 1300–11, doi:10.1093/molbev/msm049, PMID 17351267 Also see Supplementary Data
- ↑ Evatt, Danny (2013-11-01). "The Evatt Clan: A Worldwide Historical Review of the Evatt Family Surname". https://books.google.com/books?id=3QYRAgAAQBAJ&q=r1a1+age&pg=PT7.
- ↑ Sjödin, Per; François, Olivier (2011). "Wave-of-Advance Models of the Diffusion of the y Chromosome Haplogroup R1b1b2 in Europe". PLOS ONE 6 (6): e21592. doi:10.1371/journal.pone.0021592. PMID 21720564. Bibcode: 2011PLoSO...621592S.
- ↑ 53.0 53.1 Underhill, Peter A.; Myres, Natalie M.; Rootsi, Siiri; Metspalu, Mait; Zhivotovsky, Lev A.; King, Roy J.; Lin, Alice A.; Chow, Cheryl-Emiliane T. et al. (2010). "Separating the post-Glacial coancestry of European and Asian y chromosomes within haplogroup R1a". European Journal of Human Genetics 18 (4): 479–84. doi:10.1038/ejhg.2009.194. PMID 19888303.
- ^ 2005 Y-chromosome Phylogenetic Tree, from FamilyTreeDNA.com
- ^ A Nomenclature system for the Tree of Human Y-Chromosomal Haplogroups, Genome.org
Further reading
- Mendez, Fernando; Krahn, Thomas; Schrack, Bonnie; Krahn, Astrid-Maria; Veeramah, Krishna; Woerner, August; Fomine, Forka Leypey Mathew; Bradman, Neil et al. (7 March 2013). "An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree". American Journal of Human Genetics 92 (3): 454–59. doi:10.1016/j.ajhg.2013.02.002. PMID 23453668. PMC 3591855. http://haplogroup-a.com/Ancient-Root-AJHG2013.pdf.
- "Y-Haplogroup A Phylogenetic Tree". March 2013. http://www.haplogroup-a.com/A-Phylotree.html. (chart highlighting new branches added to the A phylotree in March 2013)
External links
- ISOGG Y-DNA Haplogroup Tree
- Family Tree DNA Public Haplotree
- Chart of the speed of different Y chromosomal STR mutation rates
- Map of Y Haplogroups
- Atlas of the Human Journey, from the Genographic Project, National Geographic
- DNA Heritage's Y-haplogroup map
- Video tutorial on Discovering Paternal Ancestry with Y-Chromosomes
- Haplogroup Predictor
- "The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective". Science 290 (5494): 1155–59. November 2000. doi:10.1126/science.290.5494.1155. PMID 11073453. Bibcode: 2000Sci...290.1155S. As PDF Paper that defined "Eu" haplogroups
- Y-DNA Haplogroup and Sub-clade Projects
- Kerchner's YDNA Haplogroup Descriptions, Projects & Links
- Y-DNA Testing Company STR Marker Comparison Chart
- Y-DNA Ethnographic and Genographic Atlas and Open-Source Data Compilation
- Y Chromosome Consortium
 |



