Biology:FolE RNA motif
| folE | |
|---|---|
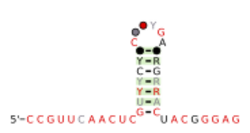 Consensus secondary structure and sequence conservation of folE RNA | |
| Identifiers | |
| Symbol | folE |
| Rfam | RF02977 |
| Other data | |
| RNA type | Cis-reg |
| SO | 0005836 |
| PDB structures | PDBe |
The folE RNA motif, now known as the THF-II riboswitch, is a conserved RNA structure that was discovered by bioinformatics.[1] folE motifs are found in Alphaproteobacteria.
folE motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. Instances of the folE RNA motif are often located nearby to the predicted Shine-Dalgarno sequence of the downstream gene. This arrangement is consistent with a model of cis-regulation where the RNA allosterically controls access to the Shine-Dalgarno sequence, thus regulating the gene translationally. All known folE RNAs are present upstream of genes encoding GTP cyclohydrolase I, which performs a step in folate metabolism.
folE RNAs have been shown to bind tetrahydrofolate and related molecules, leading to their designation as a second structural class of tetrahydrofolate riboswitches, called THF-II riboswitches.[2]
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
- ↑ "Biochemical validation of a second class of tetrahydrofolate riboswitches in bacteria". RNA 25 (9): 1091–1097. September 2019. doi:10.1261/rna.071829.119. PMID 31186369.
 |

