Biology:Bridged nucleic acid
A bridged nucleic acid (BNA) is a modified RNA nucleotide. They are sometimes also referred to as constrained or inaccessible RNA molecules. BNA monomers can contain a five-membered, six-membered or even a seven-membered bridged structure with a "fixed" C3'-endo sugar puckering.[1] The bridge is synthetically incorporated at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoramidite chemistry. BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability.
Chemical structures
Chemical structures of BNA monomers containing a bridge at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer as synthesized by Takeshi Imanishi's group.[2][3][4][5][6][7] The nature of the bridge can vary for different types of monomers. The 3D structures for A-RNA and B-DNA were used as a template for the design of the BNA monomers. The goal for the design was to find derivatives that possess high binding affinities with complementary RNA and/or DNA strands.
An increased conformational inflexibility of the sugar moiety in nucleosides (oligonucleotides) results in a gain of high binding affinity with complementary single-stranded RNA and/or double-stranded DNA. The first 2',4'-BNA (LNA) monomers were first synthesized by Takeshi Imanishi's group in 1997[2] followed independently by Jesper Wengel's group in 1998.[8]
BNA nucleotides can be incorporated into DNA or RNA oligonucleotides at any desired position. Such oligomers are synthesized chemically and are now commercially available. The bridged ribose conformation enhances base stacking and pre-organizes the backbone of the oligonucleotide significantly increasing their hybridization properties.
The incorporation of BNAs into oligonucleotides allows the production of modified synthetic oligonucleotides with equal or higher binding affinity against a DNA or RNA complement with excellent single-mismatch discriminating power; better RNA selective binding; stronger and more sequence selective triplex-forming characters; pronounced higher nuclease resistance, even higher than Sp-phosphorothioate analogues; and good aqueous solubility of the resulting oligonucleotides when compared to regular DNA or RNA oligonucleotides.
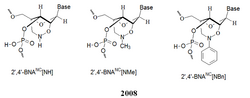
New BNA analogs introduced by Imanishi's group were designed by taking the length of the bridged moiety into account. A six-membered bridged structure with a unique structural feature (N-O bond) in the sugar moiety was designed to have a nitrogen atom. This atom improves the formation of duplexes and triplexes by lowering the repulsion between the negatively charged backbone phosphates. These modifications allow to control the affinity towards complementary strands, regulate resistance against nuclease degradation and the synthesis of functional molecules designed for specific applications in genomics. The properties of these analogs were investigated and compared to those of previous 2',4'-BNA (LNA) modified oligonucleotides by Imanishi's group. Imanishi's results show that "2',4'-BNANC-modified oligonucleotides with these profiles show great promise for applications in antisense and antigene technologies."
Proposed mechanism of action of AONs
Yamamoto et al. in 2012[9] demonstrated that BNA-based antisense therapeutics inhibited hepatic PCSK9 expression, resulting in a strong reduction of the serum LDL-C levels of mice. The findings supported the hypothesis that PCSK9 is a potential therapeutic target for hypercholesterolemia and the researchers were able to show that BNA-based antisense oligonucleotides (AONs) induced cholesterol-lowering action in hypercholesterolemic mice. A moderate increase of aspartate aminotransferase, ALT, and blood urea nitrogen levels was observed whereas the histopathological analysis revealed no severe hepatic toxicities. The same group, also in 2012, reported that the 2',4'-BNANC[NMe] analog when used in antisense oligonucleotides showed significantly stronger inhibitory activities which is more pronounced in shorter (13- to 16mer) oligonucleotides. Their data led the researchers to conclude that the 2',4'-BNANC[NMe] analog may be a better alternative to conventional LNAs.
Benefits of the BNA technology
Some of the benefits of BNAs include ideal for the detection of short RNA and DNA targets; increase the thermal stability of duplexes; capable of single nucleotide discrimination; increases the thermal stability of triplexes; resistance to exo- and endonucleases resulting in a high stability for in vivo and in vitro applications; increased target specificity; facilitate Tm normalization; strand invasion enables detection of "hard to access" samples; compatible with standard enzymatic processes.[citation needed]
Application of the BNA technology
Application of BNAs include small RNA research; design and synthesis of RNA aptamers; siRNA; antisense probes; diagnostics; isolation; microarray analysis; Northern blotting; real-time PCR; in situ hybridization; functional analysis; SNP detection and use as antigens and many others nucleotide base applications.[10][citation needed]
References
- ↑ Saenger, W. (1984) Principles of Nucleic Acid Structure, Springer-Verlag, New York, ISBN:3-540-90761-0.
- ↑ 2.0 2.1 Obika, S.; Nanbu, D.; Hari, Y.; Morio, K. I.; In, Y.; Ishida, T.; Imanishi, T. (1997). "Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering". Tetrahedron Letters 38 (50): 8735. doi:10.1016/S0040-4039(97)10322-7.
- ↑ Obika, S.; Onoda, M.; Andoh, K.; Imanishi, J.; Morita, M.; Koizumi, T. (2001). "3'-amino-2',4'-BNA: Novel bridged nucleic acids having an N3'-->P5' phosphoramidate linkage". Chemical Communications (19): 1992–1993. doi:10.1039/b105640a. PMID 12240255.
- ↑ Obika, Satoshi; Hari, Yoshiyuki; Sekiguchi, Mitsuaki; Imanishi, Takeshi (2001). "A 2′,4′-Bridged Nucleic Acid Containing 2-Pyridone as a Nucleobase: Efficient Recognition of a C⋅G Interruption by Triplex Formation with a Pyrimidine Motif". Angewandte Chemie International Edition 40 (11): 2079. doi:10.1002/1521-3773(20010601)40:11<2079::AID-ANIE2079>3.0.CO;2-Z.
- ↑ Morita, K.; Hasegawa, C.; Kaneko, M.; Tsutsumi, S.; Sone, J.; Ishikawa, T.; Imanishi, T.; Koizumi, M. (2001). "2'-O,4'-C-ethylene-bridged nucleic acids (ENA) with nuclease-resistance and high affinity for RNA". Nucleic Acids Research. Supplement 1 (1): 241–242. doi:10.1093/nass/1.1.241. PMID 12836354.
- ↑ Hari, Y.; Obika, S.; Sekiguchi, M.; Imanishi, T. (2003). "Selective recognition of CG interruption by 2′,4′-BNA having 1-isoquinolone as a nucleobase in a pyrimidine motif triplex formation". Tetrahedron 59 (27): 5123. doi:10.1016/S0040-4020(03)00728-2.
- ↑ 7.0 7.1 Rahman, S. M. A.; Seki, S.; Obika, S.; Haitani, S.; Miyashita, K.; Imanishi, T. (2007). "Highly Stable Pyrimidine-Motif Triplex Formation at Physiological pH Values by a Bridged Nucleic Acid Analogue". Angewandte Chemie International Edition 46 (23): 4306–4309. doi:10.1002/anie.200604857. PMID 17469090.
- ↑ Koshkin, A. A.; Singh, S. K.; Nielsen, P.; Rajwanshi, V. K.; Kumar, R.; Meldgaard, M.; Olsen, C. E.; Wengel, J. (1998). "LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition". Tetrahedron 54 (14): 3607. doi:10.1016/S0040-4020(98)00094-5.
- ↑ Koizumi, M. (2006). "ENA oligonucleotides as therapeutics". Current Opinion in Molecular Therapeutics 8 (2): 144–149. PMID 16610767.
- ↑ Soler-Bistué, Alfonso; Zorreguieta, Angeles; Tolmasky, Marcelo E. (31 May 2019). "Bridged Nucleic Acids Reloaded" (in en). Molecules 24 (12): 2297. doi:10.3390/molecules24122297. PMID 31234313.
External links
- https://web.archive.org/web/20130126055902/http://www.rockefeller.edu/labheads/tuschl/sirna.html
- http://www.sanger.ac.uk/resources/software/
- Pfundheller, Henrik M.; Lomholt, Christian (2002). "Locked Nucleic Acids: Synthesis and Characterization of LNA-T Diol". Locked Nucleic Acids: Synthesis and Characterization of LNA‐T Diol. Chapter 4. 4.12.1–4.12.16. doi:10.1002/0471142700.nc0412s08. ISBN 978-0471142706.
 |





