Biology:Amine oxidase (copper-containing)
| amine oxidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.4.3.6 | ||||||||
| CAS number | 9001-53-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Copper amine oxidase, enzyme domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
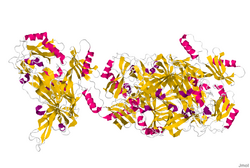 Crystal structure of a copper-containing benzylamine oxidase from Hansenula polymorpha.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Cu_amine_oxid | ||||||||
| Pfam | PF01179 | ||||||||
| InterPro | IPR015798 | ||||||||
| PROSITE | PDOC00895 | ||||||||
| SCOP2 | 1oac / SCOPe / SUPFAM | ||||||||
| Membranome | 252 | ||||||||
| |||||||||
| Copper amine oxidase N-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
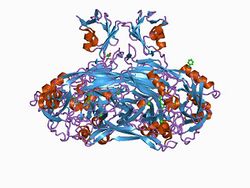 crystal structure of e. coli amine oxidase anaerobically reduced with beta-phenylethylamine | |||||||||
| Identifiers | |||||||||
| Symbol | Cu_amine_oxidN1 | ||||||||
| Pfam | PF07833 | ||||||||
| InterPro | IPR012854 | ||||||||
| SCOP2 | 1spu / SCOPe / SUPFAM | ||||||||
| |||||||||
| Copper amine oxidase, N2 domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2a resolution | |||||||||
| Identifiers | |||||||||
| Symbol | Cu_amine_oxidN2 | ||||||||
| Pfam | PF02727 | ||||||||
| Pfam clan | CL0047 | ||||||||
| InterPro | IPR015800 | ||||||||
| PROSITE | PDOC00895 | ||||||||
| SCOP2 | 1oac / SCOPe / SUPFAM | ||||||||
| |||||||||
| Copper amine oxidase, N3 domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of hansenula polymorpha amine oxidase in complex with xe to 1.6 angstroms | |||||||||
| Identifiers | |||||||||
| Symbol | Cu_amine_oxidN3 | ||||||||
| Pfam | PF02728 | ||||||||
| Pfam clan | CL0047 | ||||||||
| InterPro | IPR015802 | ||||||||
| PROSITE | PDOC00895 | ||||||||
| SCOP2 | 1oac / SCOPe / SUPFAM | ||||||||
| |||||||||
Amine oxidase (copper-containing) (AOC) (EC 1.4.3.21 and EC 1.4.3.22; formerly EC 1.4.3.6) is a family of amine oxidase enzymes which includes both primary-amine oxidase and diamine oxidase; these enzymes catalyze the oxidation of a wide range of biogenic amines including many neurotransmitters, histamine and xenobiotic amines. They act as a disulphide-linked homodimer. They catalyse the oxidation of primary amines to aldehydes, with the subsequent release of ammonia and hydrogen peroxide, which requires one copper ion per subunit and topaquinone as cofactor:[2]
- RCH2NH2 + H2O + O2 [math]\displaystyle{ \rightleftharpoons }[/math] RCHO + NH3 + H2O2
The 3 substrates of this enzyme are primary amines (RCH2NH2), H2O, and O2, whereas its 3 products are RCHO, NH3, and H2O2.
Copper-containing amine oxidases are found in bacteria, fungi, plants and animals. In prokaryotes, the enzyme enables various amine substrates to be used as sources of carbon and nitrogen.[3][4]
This enzyme belongs to oxidoreductases, specifically those acting on the CH-NH2 group of donors with oxygen as acceptor. The systematic name of this enzyme class is amine:oxygen oxidoreductase (deaminating) (copper-containing). This enzyme participates in 8 metabolic pathways: urea cycle and metabolism of amino groups, glycine, serine and threonine metabolism, histidine metabolism, tyrosine metabolism, phenylalanine metabolism, tryptophan metabolism, beta-alanine metabolism, and alkaloid biosynthesis ii. It has 2 cofactors: copper, and PQQ.
Structure
The copper amine oxidase 3-dimensional structure was determined through X-ray crystallography.[1] The copper amine oxidases occur as mushroom-shaped homodimers of 70-95 kDa, each monomer containing a copper ion and a covalently bound redox cofactor, topaquinone (TPQ). TPQ is formed by post-translational modification of a conserved tyrosine residue. The copper ion is coordinated with three histidine residues and two water molecules in a distorted square pyramidal geometry, and has a dual function in catalysis and TPQ biogenesis. The catalytic domain is the largest of the 3-4 domains found in copper amine oxidases, and consists of a beta sandwich of 18 strands in two sheets. The active site is buried and requires a conformational change to allow the substrate access.
The N2 and N3 N-terminal domains share a common structural fold, its core consisting of alpha-beta(4), where the helix is packed against the coiled anti-parallel beta-sheets. An additional domain is found at the N-terminal of some copper amine oxidases, as well as in related proteins such as cell wall hydrolase and N-acetylmuramoyl-L-alanine amidase. This domain consists of a five-stranded antiparallel beta-sheet twisted around an alpha helix.[5][6]
Function
In eukaryotes they have a broader range of functions, including cell differentiation and growth, wound healing, detoxification and cell signalling;[7] one AOC enzyme (AOC3) functions as a vascular adhesion protein (VAP-1) in some mammalian tissues.[1]
Human proteins containing this domain
See also
- Copper in health
References
- ↑ 1.0 1.1 1.2 PDB: 3LOY; "Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha". Biochemistry 49 (11): 2540–50. March 2010. doi:10.1021/bi901933d. PMID 20155950.
- ↑ "Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution". Structure 3 (11): 1171–1184. 1995. doi:10.1016/s0969-2126(01)00253-2. PMID 8591028.
- ↑ "Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction". Biochemistry 36 (7): 1608–1620. 1997. doi:10.1021/bi962205j. PMID 9048544.
- ↑ "Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone". Biochemistry 36 (51): 16116–16133. 1997. doi:10.1021/bi971797i. PMID 9405045.
- ↑ "Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution". Structure 3 (11): 1171–84. November 1995. doi:10.1016/s0969-2126(01)00253-2. PMID 8591028.
- ↑ "Visualization of dioxygen bound to copper during enzyme catalysis". Science 286 (5445): 1724–8. November 1999. doi:10.1126/science.286.5445.1724. PMID 10576737.
- ↑ "Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution". Structure 4 (8): 943–955. 1996. doi:10.1016/s0969-2126(96)00101-3. PMID 8805580.
Further reading
- "Microbial-production of pyrroloquinoline quinone". Agric. Biol. Chem. 48 (2): 561–565. 1984. doi:10.1271/bbb1961.48.561.
- "Esterases in the milk and blood plasma of swine. I. Substrate specificity and electrophoresis studies". Biochem. J. 71 (3): 477–84. 1959. doi:10.1042/bj0710477. PMID 13638253.
- Boyer, P.D., Lardy, H. and Myrback, K. (Eds.), The Enzymes, 2nd ed., vol. 8, Academic Press, New York, 1963, p. 337-351.
- "Benzylamine oxidase and histaminase: purification and crystallization of an enzyme from pig plasma". Proceedings of the Royal Society B 161 (983): 153–67. 1964. doi:10.1098/rspb.1964.0086. PMID 14224405. Bibcode: 1964RSPSB.161..153B.
- "Microbial oxidation of amines. Distribution, purification and properties of two primary-amine oxidases from the yeast Candida boidinii grown on amines as sole nitrogen source". Biochem. J. 199 (1): 187–201. 1981. doi:10.1042/bj1990187. PMID 7337701.
- McEwen CM Jr (1965). "Human plasma monoamine oxidase. 1. Purification and identification". J. Biol. Chem. 240 (5): 2003–10. PMID 5888801.
- "Pyridoxal phosphate as a prosthetic group of pig kidney diamine oxidase". Arch. Biochem. Biophys. 119 (1): 373–81. 1967. doi:10.1016/0003-9861(67)90468-7. PMID 4964016.
- "Amine oxidases of microorganisms. Part II. Purification and crystallisation of amine oxidase of Aspergillus niger". Agric. Biol. Chem. 29: 649–654. 1965. doi:10.1271/bbb1961.29.649.
- "Amine oxidases of microorganisms. Part III. Properties of amine oxidase of Aspergillus niger". Agric. Biol. Chem. 29: 864–869. 1965. doi:10.1271/bbb1961.29.864.
- "Amine oxidases of microorganisms. Part IV. Further properties of amine oxidase of Aspergillus niger". Agric. Biol. Chem. 29: 912–917. 1965. doi:10.1271/bbb1961.29.912.
- Boyer, P.D., Lardy, H. and Myrback, K. (Eds.), The Enzymes, 2nd ed., vol. 8, Academic Press, New York, 1963, p. 313-335.
 |

